Survey of the infant male urobiome and genomic analysis of Actinotignum spp
- PMID: 38040700
- PMCID: PMC10692110
- DOI: 10.1038/s41522-023-00457-6
Survey of the infant male urobiome and genomic analysis of Actinotignum spp
Abstract
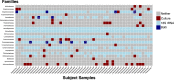
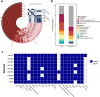
The urinary bladder harbors a community of microbes termed the urobiome, which remains understudied. In this study, we present the urobiome of healthy infant males from samples collected by transurethral catheterization. Using a combination of enhanced culture and amplicon sequencing, we identify several common bacterial genera that can be further investigated for their effects on urinary health across the lifespan. Many genera were shared between all samples suggesting a consistent urobiome composition among this cohort. We note that, for this cohort, early life exposures including mode of birth (vaginal vs. Cesarean section), or prior antibiotic exposure did not influence urobiome composition. In addition, we report the isolation of culturable bacteria from the bladders of these infant males, including Actinotignum spp., a bacterial genus that has been associated with urinary tract infections in older male adults. Herein, we isolate and sequence 9 distinct strains of Actinotignum spp. enhancing the genomic knowledge surrounding this genus and opening avenues for delineating the microbiology of this urobiome constituent. Furthermore, we present a framework for using the combination of culture-dependent and sequencing methodologies for uncovering mechanisms in the urobiome.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Update of
-
Defining the Infant Male Urobiome and Moving Towards Mechanisms in Urobiome Research.Res Sq [Preprint]. 2023 Mar 7:rs.3.rs-2618137. doi: 10.21203/rs.3.rs-2618137/v1. Res Sq. 2023. Update in: NPJ Biofilms Microbiomes. 2023 Dec 1;9(1):91. doi: 10.1038/s41522-023-00457-6. PMID: 36945625 Free PMC article. Updated. Preprint.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

