hsa_circ_0007919 induces LIG1 transcription by binding to FOXA1/TET1 to enhance the DNA damage response and promote gemcitabine resistance in pancreatic ductal adenocarcinoma
- PMID: 38044421
- PMCID: PMC10694898
- DOI: 10.1186/s12943-023-01887-8
hsa_circ_0007919 induces LIG1 transcription by binding to FOXA1/TET1 to enhance the DNA damage response and promote gemcitabine resistance in pancreatic ductal adenocarcinoma
Erratum in
-
Correction: hsa_circ_0007919 induces LIG1 transcription by binding to FOXA1/TET1 to enhance the DNA damage response and promote gemcitabine resistance in pancreatic ductal adenocarcinoma.Mol Cancer. 2024 Jan 16;23(1):16. doi: 10.1186/s12943-024-01937-9. Mol Cancer. 2024. PMID: 38229124 Free PMC article. No abstract available.
Abstract
Background: Circular RNAs (circRNAs) play important roles in the occurrence and development of cancer and chemoresistance. DNA damage repair contributes to the proliferation of cancer cells and resistance to chemotherapy-induced apoptosis. However, the role of circRNAs in the regulation of DNA damage repair needs clarification.
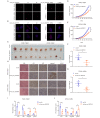
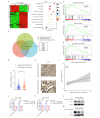
Methods: RNA sequencing analysis was applied to identify the differentially expressed circRNAs. qRT-PCR was conducted to confirm the expression of hsa_circ_0007919, and CCK-8, FCM, single-cell gel electrophoresis and IF assays were used to analyze the proliferation, apoptosis and gemcitabine (GEM) resistance of pancreatic ductal adenocarcinoma (PDAC) cells. Xenograft model and IHC experiments were conducted to confirm the effects of hsa_circ_0007919 on tumor growth and DNA damage in vivo. RNA sequencing and GSEA were applied to confirm the downstream genes and pathways of hsa_circ_0007919. FISH and nuclear-cytoplasmic RNA fractionation experiments were conducted to identify the cellular localization of hsa_circ_0007919. ChIRP, RIP, Co-IP, ChIP, MS-PCR and luciferase reporter assays were conducted to confirm the interaction among hsa_circ_0007919, FOXA1, TET1 and the LIG1 promoter.
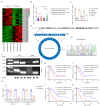
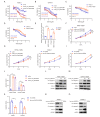
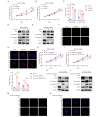
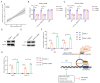
Results: We identified a highly expressed circRNA, hsa_circ_0007919, in GEM-resistant PDAC tissues and cells. High expression of hsa_circ_0007919 correlates with poor overall survival (OS) and disease-free survival (DFS) of PDAC patients. Hsa_circ_0007919 inhibits the DNA damage, accumulation of DNA breaks and apoptosis induced by GEM in a LIG1-dependent manner to maintain cell survival. Mechanistically, hsa_circ_0007919 recruits FOXA1 and TET1 to decrease the methylation of the LIG1 promoter and increase its transcription, further promoting base excision repair, mismatch repair and nucleotide excision repair. At last, we found that GEM enhanced the binding of QKI to the introns of hsa_circ_0007919 pre-mRNA and the splicing and circularization of this pre-mRNA to generate hsa_circ_0007919.
Conclusions: Hsa_circ_0007919 promotes GEM resistance by enhancing DNA damage repair in a LIG1-dependent manner to maintain cell survival. Targeting hsa_circ_0007919 and DNA damage repair pathways could be a therapeutic strategy for PDAC.
Keywords: DNA damage repair; LIG1; Pancreatic ductal adenocarcinoma; QKI; hsa_circ_0007919.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no potential conflicts of interest.
Figures


Similar articles
-
Circular RNA circ-MTHFD1L induces HR repair to promote gemcitabine resistance via the miR-615-3p/RPN6 axis in pancreatic ductal adenocarcinoma.J Exp Clin Cancer Res. 2022 Apr 23;41(1):153. doi: 10.1186/s13046-022-02343-z. J Exp Clin Cancer Res. 2022. PMID: 35459186 Free PMC article.
-
Circular RNA hsa_circ_0007367 promotes the progression of pancreatic ductal adenocarcinoma by sponging miR-6820-3p and upregulating YAP1 expression.Cell Death Dis. 2022 Aug 25;13(8):736. doi: 10.1038/s41419-022-05188-8. Cell Death Dis. 2022. PMID: 36008392 Free PMC article.
-
Upregulated circular RNA circ_0007534 indicates an unfavorable prognosis in pancreatic ductal adenocarcinoma and regulates cell proliferation, apoptosis, and invasion by sponging miR-625 and miR-892b.J Cell Biochem. 2019 Mar;120(3):3780-3789. doi: 10.1002/jcb.27658. Epub 2018 Nov 1. J Cell Biochem. 2019. PMID: 30382592
-
Mobius strip in pancreatic cancer: biogenesis, function and clinical significance of circular RNAs.Cell Mol Life Sci. 2021 Sep;78(17-18):6201-6213. doi: 10.1007/s00018-021-03908-5. Epub 2021 Aug 3. Cell Mol Life Sci. 2021. PMID: 34342664 Free PMC article. Review.
-
Prognostic and clinicopathological roles of circular RNA expression in chemoresistance in head and neck squamous cell carcinoma: a systematic review.Front Pharmacol. 2025 Mar 19;16:1502107. doi: 10.3389/fphar.2025.1502107. eCollection 2025. Front Pharmacol. 2025. PMID: 40176914 Free PMC article.
Cited by
-
The crosstalk between alternative splicing and circular RNA in cancer: pathogenic insights and therapeutic implications.Cell Mol Biol Lett. 2024 Nov 16;29(1):142. doi: 10.1186/s11658-024-00662-x. Cell Mol Biol Lett. 2024. PMID: 39550559 Free PMC article. Review.
-
Role of circular RNAs in DNA repair.RNA Biol. 2024 Jan;21(1):149-161. doi: 10.1080/15476286.2024.2429945. Epub 2024 Nov 17. RNA Biol. 2024. PMID: 39550713 Free PMC article. Review.
-
Niraparib perturbs autophagosome-lysosome fusion in pancreatic ductal adenocarcinoma and exhibits anticancer potential against gemcitabine-resistant PDAC.Transl Oncol. 2025 Jan;51:102206. doi: 10.1016/j.tranon.2024.102206. Epub 2024 Nov 27. Transl Oncol. 2025. PMID: 39603206 Free PMC article.
-
Development of a Nomogram Integrating Modified Inflammation-Based Indexes for Predicting Overall Survival in Pancreatic Cancer: A Retrospective Study.J Inflamm Res. 2025 Apr 7;18:4813-4830. doi: 10.2147/JIR.S519779. eCollection 2025. J Inflamm Res. 2025. PMID: 40224394 Free PMC article.
-
Circular RNAs in cancer.MedComm (2020). 2025 Feb 2;6(2):e70079. doi: 10.1002/mco2.70079. eCollection 2025 Feb. MedComm (2020). 2025. PMID: 39901896 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

