This is a preprint.
Multimodal Biomedical Data Fusion Using Sparse Canonical Correlation Analysis and Cooperative Learning: A Cohort Study on COVID-19
- PMID: 38045288
- PMCID: PMC10690316
- DOI: 10.21203/rs.3.rs-3569833/v1
Multimodal Biomedical Data Fusion Using Sparse Canonical Correlation Analysis and Cooperative Learning: A Cohort Study on COVID-19
Update in
-
Multimodal data fusion using sparse canonical correlation analysis and cooperative learning: a COVID-19 cohort study.NPJ Digit Med. 2024 May 7;7(1):117. doi: 10.1038/s41746-024-01128-2. NPJ Digit Med. 2024. PMID: 38714751 Free PMC article.
Abstract
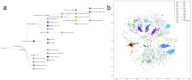
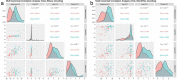
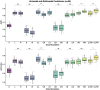
Through technological innovations, patient cohorts can be examined from multiple views with high-dimensional, multiscale biomedical data to classify clinical phenotypes and predict outcomes. Here, we aim to present our approach for analyzing multimodal data using unsupervised and supervised sparse linear methods in a COVID-19 patient cohort. This prospective cohort study of 149 adult patients was conducted in a tertiary care academic center. First, we used sparse canonical correlation analysis (CCA) to identify and quantify relationships across different data modalities, including viral genome sequencing, imaging, clinical data, and laboratory results. Then, we used cooperative learning to predict the clinical outcome of COVID-19 patients. We show that serum biomarkers representing severe disease and acute phase response correlate with original and wavelet radiomics features in the LLL frequency channel (corr(Xu1, Zv1) = 0.596, p-value < 0.001). Among radiomics features, histogram-based first-order features reporting the skewness, kurtosis, and uniformity have the lowest negative, whereas entropy-related features have the highest positive coefficients. Moreover, unsupervised analysis of clinical data and laboratory results gives insights into distinct clinical phenotypes. Leveraging the availability of global viral genome databases, we demonstrate that the Word2Vec natural language processing model can be used for viral genome encoding. It not only separates major SARS-CoV-2 variants but also allows the preservation of phylogenetic relationships among them. Our quadruple model using Word2Vec encoding achieves better prediction results in the supervised task. The model yields area under the curve (AUC) and accuracy values of 0.87 and 0.77, respectively. Our study illustrates that sparse CCA analysis and cooperative learning are powerful techniques for handling high-dimensional, multimodal data to investigate multivariate associations in unsupervised and supervised tasks.
Figures



References
-
- Topol E.J. High-performance medicine: the convergence of human and artificial intelligence. Nature Medicine 25, 44–56 (2019). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

