This is a preprint.
Distinct evolution of SARS-CoV-2 Omicron XBB and BA.2.86/JN.1 lineages combining increased fitness and antibody evasion
- PMID: 38045308
- PMCID: PMC10690205
- DOI: 10.1101/2023.11.20.567873
Distinct evolution of SARS-CoV-2 Omicron XBB and BA.2.86/JN.1 lineages combining increased fitness and antibody evasion
Update in
-
Distinct evolution of SARS-CoV-2 Omicron XBB and BA.2.86/JN.1 lineages combining increased fitness and antibody evasion.Nat Commun. 2024 Mar 13;15(1):2254. doi: 10.1038/s41467-024-46490-7. Nat Commun. 2024. PMID: 38480689 Free PMC article.
Abstract
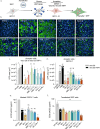
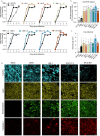
The unceasing circulation of SARS-CoV-2 leads to the continuous emergence of novel viral sublineages. Here, we isolated and characterized XBB.1, XBB.1.5, XBB.1.9.1, XBB.1.16.1, EG.5.1.1, EG.5.1.3, XBF, BA.2.86.1 and JN.1 variants, representing >80% of circulating variants in January 2024. The XBB subvariants carry few but recurrent mutations in the spike, whereas BA.2.86.1 and JN.1 harbor >30 additional changes. These variants replicated in IGROV-1 but no longer in Vero E6 and were not markedly fusogenic. They potently infected nasal epithelial cells, with EG.5.1.3 exhibiting the highest fitness. Antivirals remained active. Neutralizing antibody (NAb) responses from vaccinees and BA.1/BA.2-infected individuals were markedly lower compared to BA.1, without major differences between variants. An XBB breakthrough infection enhanced NAb responses against both XBB and BA.2.86 variants. JN.1 displayed lower affinity to ACE2 and higher immune evasion properties compared to BA.2.86.1. Thus, while distinct, the evolutionary trajectory of these variants combines increased fitness and antibody evasion.
Conflict of interest statement
Competing Interests Statement The authors declare no competing interests.
Figures





References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
