This is a preprint.
A Potent Kalihinol Analogue Disrupts Apicoplast Function and Vesicular Trafficking in P. falciparum Malaria
- PMID: 38045341
- PMCID: PMC10690269
- DOI: 10.1101/2023.11.21.568162
A Potent Kalihinol Analogue Disrupts Apicoplast Function and Vesicular Trafficking in P. falciparum Malaria
Update in
-
A kalihinol analog disrupts apicoplast function and vesicular trafficking in P. falciparum malaria.Science. 2024 Sep 27;385(6716):eadm7966. doi: 10.1126/science.adm7966. Epub 2024 Sep 27. Science. 2024. PMID: 39325875 Free PMC article.
Abstract
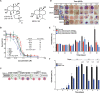
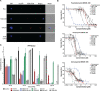
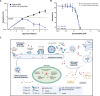
Here we report the discovery of MED6-189, a new analogue of the kalihinol family of isocyanoterpene (ICT) natural products. MED6-189 is effective against drug-sensitive and -resistant P. falciparum strains blocking both intraerythrocytic asexual replication and sexual differentiation. This compound was also effective against P. knowlesi and P. cynomolgi. In vivo efficacy studies using a humanized mouse model of malaria confirms strong efficacy of the compound in animals with no apparent hemolytic activity or apparent toxicity. Complementary chemical biology, molecular biology, genomics and cell biological analyses revealed that MED6-189 primarily targets the parasite apicoplast and acts by inhibiting lipid biogenesis and cellular trafficking. Genetic analyses in P. falciparum revealed that a mutation in PfSec13, which encodes a component of the parasite secretory machinery, reduced susceptibility to the drug. The high potency of MED6-189 in vitro and in vivo, its broad range of efficacy, excellent therapeutic profile, and unique mode of action make it an excellent addition to the antimalarial drug pipeline.
Conflict of interest statement
Competing Interests-- The authors declare no competing interests. Correspondence and requests for materials should be addressed to Karine Le Roch.
Figures


References
-
- WHO, World malaria report 2022. (2022).
-
- Organization W. H., Global report on antimalarial drug efficacy and drug resistance, 2000–2010. WHO, (2010).
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
