Structome: a tool for the rapid assembly of datasets for structural phylogenetics
- PMID: 38046099
- PMCID: PMC10692761
- DOI: 10.1093/bioadv/vbad134
Structome: a tool for the rapid assembly of datasets for structural phylogenetics
Abstract
Summary: Protein structures carry signal of common ancestry and can therefore aid in reconstructing their evolutionary histories. To expedite the structure-informed inference process, a web server, Structome, has been developed that allows users to rapidly identify protein structures similar to a query protein and to assemble datasets useful for structure-based phylogenetics. Structome was created by clustering of the structures in RCSB PDB using 90% sequence identity and representing each cluster by a centroid structure. Structure similarity between centroid proteins was calculated, and annotations from PDB, SCOP, and CATH were integrated. To illustrate utility, an H3 histone was used as a query, and results show that the protein structures returned by Structome span both sequence and structural diversity of the histone fold. Additionally, the pre-computed nexus-formatted distance matrix, provided by Structome, enables analysis of evolutionary relationships between proteins not identifiable using searches based on sequence similarity alone. Our results demonstrate that, beginning with a single structure, Structome can be used to rapidly generate a dataset of structural neighbours and allows deep evolutionary history of proteins to be studied.
Availability and implementation: Structome is available at: https://structome.bii.a-star.edu.sg.
© The Author(s) 2023. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures
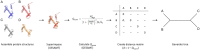



References
LinkOut - more resources
Full Text Sources

