pH Dependent Reversible Formation of a Binuclear Ni2 Metal-Center Within a Peptide Scaffold
- PMID: 38046130
- PMCID: PMC10691859
- DOI: 10.3390/inorganics7070090
pH Dependent Reversible Formation of a Binuclear Ni2 Metal-Center Within a Peptide Scaffold
Abstract
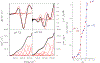
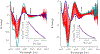
A disulfide-bridged peptide containing two Ni2+ binding sites based on the nickel superoxide dismutase protein, {Ni2(SODmds)}, has been prepared. At physiological pH (7.4) it was found that the metal sites are mononuclear with a square planar NOS2 coordination environment with the two sulfur-based ligands derived from cysteinate residues, the nitrogen ligand derived from the amide backbone and a water ligand. Furthermore, S K-edge X-ray absorption spectroscopy indicated that the two cysteinate sulfur atoms ligated to nickel are each protonated. Elevation of the pH to 9.6 results in the deprotonation of the cysteinate sulfur atoms, and yields a binuclear, cysteinate bridged Ni22+ center with each nickel contained in a distorted square planar geometry. At both pH = 7.4 and 9.6 the nickel sites are moderately air sensitive, yielding intractable oxidation products. However, at pH = 9.6 {Ni2(SODmds)} reacts with O2 at an ~3.5-fold faster rate than at pH = 7.4. Electronic structure calculations indicate the reduced reactivity at pH = 7.4 is a result of a reduction in S(3p) character and deactivation of the nucleophilic frontier molecular orbitals upon cysteinate sulfur protonation.
Keywords: biological nickel sites; dinuclear nickel metallopeptides; nickel-thiolates; thiolate oxidative damage.
Conflict of interest statement
Conflicts of Interest: The authors declare no conflict of interest.
Figures
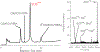
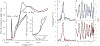


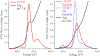
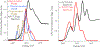


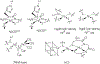

Similar articles
-
Probing variable amine/amide ligation in Ni(II)N2S2 complexes using sulfur K-edge and nickel L-edge X-ray absorption spectroscopies: implications for the active site of nickel superoxide dismutase.Inorg Chem. 2008 Apr 7;47(7):2649-60. doi: 10.1021/ic7019878. Epub 2008 Mar 11. Inorg Chem. 2008. PMID: 18330983
-
Insight into the structure and mechanism of nickel-containing superoxide dismutase derived from peptide-based mimics.Acc Chem Res. 2014 Aug 19;47(8):2332-41. doi: 10.1021/ar500060s. Epub 2014 May 13. Acc Chem Res. 2014. PMID: 24825124
-
Cysteinate protonation and water hydrogen bonding at the active-site of a nickel superoxide dismutase metallopeptide-based mimic: implications for the mechanism of superoxide reduction.J Am Chem Soc. 2014 Nov 12;136(45):16009-22. doi: 10.1021/ja5079514. Epub 2014 Nov 3. J Am Chem Soc. 2014. PMID: 25322331
-
The Role of Mixed Amine/Amide Ligation in Nickel Superoxide Dismutase.Inorg Chem. 2018 Oct 15;57(20):12521-12535. doi: 10.1021/acs.inorgchem.8b01499. Epub 2018 Oct 3. Inorg Chem. 2018. PMID: 30281299 Free PMC article.
-
The influence of amine/amide versus bisamide coordination in nickel superoxide dismutase.Inorg Chem. 2006 Dec 25;45(26):10552-66. doi: 10.1021/ic061156o. Inorg Chem. 2006. PMID: 17173410
Cited by
-
Characterization of Methyl- and Acetyl-Ni Intermediates in Acetyl CoA Synthase Formed during Anaerobic CO2 and CO Fixation.J Am Chem Soc. 2023 Jun 28;145(25):13696-13708. doi: 10.1021/jacs.3c01772. Epub 2023 Jun 12. J Am Chem Soc. 2023. PMID: 37306669 Free PMC article.
References
-
- Ragsdale SW, Biochemistry of methyl-coenzyme M reductase: the nickel metalloenzyme that catalyzes the final step in synthesis and the first step in anaerobic oxidation of the greenhouse gas methane. Met. Ions Life Sci. 2014, 14 (Metal-Driven Biogeochemistry of Gaseous Compounds in the Environment), 125–145. - PubMed
-
- Ragsdale SW, Nickel biochemistry. Curr. Opin. Chem. Biol. 1998, 2 (2), 208–215. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
