Optimizing ancestral trait reconstruction of large HIV Subtype C datasets through multiple-trait subsampling
- PMID: 38046219
- PMCID: PMC10691791
- DOI: 10.1093/ve/vead069
Optimizing ancestral trait reconstruction of large HIV Subtype C datasets through multiple-trait subsampling
Abstract
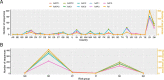
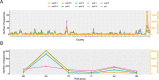
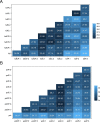
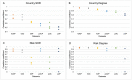
Large datasets along with sampling bias represent a challenge for phylodynamic reconstructions, particularly when the study data are obtained from various heterogeneous sources and/or through convenience sampling. In this study, we evaluate the presence of unbalanced sampled distribution by collection date, location, and risk group of human immunodeficiency virus Type 1 Subtype C using a comprehensive subsampling strategy and assess their impact on the reconstruction of the viral spatial and risk group dynamics using phylogenetic comparative methods. Our study shows that a most suitable dataset for ancestral trait reconstruction can be obtained through subsampling by all available traits, particularly using multigene datasets. We also demonstrate that sampling bias is inflated when considerable information for a given trait is unavailable or of poor quality, as we observed for the trait risk group. In conclusion, we suggest that, even if traits are not well recorded, including them deliberately optimizes the representativeness of the original dataset rather than completely excluding them. Therefore, we advise the inclusion of as many traits as possible with the aid of subsampling approaches in order to optimize the dataset for phylodynamic analysis while reducing the computational burden. This will benefit research communities investigating the evolutionary and spatio-temporal patterns of infectious diseases.
Keywords: HIV Subtype C; ancestral trait reconstruction; multiple-trait subsampling; phylogenetic comparative methods; subsampling approaches.
Published by Oxford University Press 2023. This work is written by (a) US Government employee(s) and is in the public domain in the US.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
On retrospective k-space subsampling schemes for deep MRI reconstruction.Magn Reson Imaging. 2024 Apr;107:33-46. doi: 10.1016/j.mri.2023.12.012. Epub 2024 Jan 4. Magn Reson Imaging. 2024. PMID: 38184093
-
An evaluation of phylogenetic methods for reconstructing transmitted HIV variants using longitudinal clonal HIV sequence data.J Virol. 2014 Jun;88(11):6181-94. doi: 10.1128/JVI.00483-14. Epub 2014 Mar 19. J Virol. 2014. PMID: 24648453 Free PMC article.
-
Genomic prediction using subsampling.BMC Bioinformatics. 2017 Mar 24;18(1):191. doi: 10.1186/s12859-017-1582-3. BMC Bioinformatics. 2017. PMID: 28340551 Free PMC article.
-
Toward a Functional Trait Approach to Bee Ecology.Ecol Evol. 2024 Oct 18;14(10):e70465. doi: 10.1002/ece3.70465. eCollection 2024 Oct. Ecol Evol. 2024. PMID: 39429800 Free PMC article. Review.
-
Understanding phylogenetic incongruence: lessons from phyllostomid bats.Biol Rev Camb Philos Soc. 2012 Nov;87(4):991-1024. doi: 10.1111/j.1469-185X.2012.00240.x. Epub 2012 Aug 14. Biol Rev Camb Philos Soc. 2012. PMID: 22891620 Free PMC article. Review.
Cited by
-
The emergence and circulation of human immunodeficiency virus (HIV)-1 subtype C.J Med Microbiol. 2024 May;73(5):001827. doi: 10.1099/jmm.0.001827. J Med Microbiol. 2024. PMID: 38757423 Free PMC article.
References
-
- Alzohairy A. (2011) ‘BioEdit: An Important Software for Molecular Biology’, GERF Bulletin of Biosciences, 2: 60–1.
-
- Bogdanowicz D., and Giaro K. (2012) ‘Matching Split Distance for Unrooted Binary Phylogenetic Trees’, IEEE/ACM Transactions on Computational Biology and Bioinformatics, 9: 150–60. - PubMed

