Functional and spatial proteomics profiling reveals intra- and intercellular signaling crosstalk in colorectal cancer
- PMID: 38047086
- PMCID: PMC10692669
- DOI: 10.1016/j.isci.2023.108399
Functional and spatial proteomics profiling reveals intra- and intercellular signaling crosstalk in colorectal cancer
Abstract
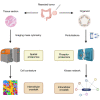
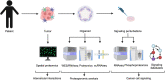
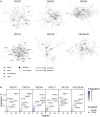
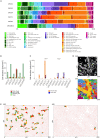
Precision oncology approaches for patients with colorectal cancer (CRC) continue to lag behind other solid cancers. Functional precision oncology-a strategy that is based on perturbing primary tumor cells from cancer patients-could provide a road forward to personalize treatment. We extend this paradigm to measuring proteome activity landscapes by acquiring quantitative phosphoproteomic data from patient-derived organoids (PDOs). We show that kinase inhibitors induce inhibitor- and patient-specific off-target effects and pathway crosstalk. Reconstruction of the kinase networks revealed that the signaling rewiring is modestly affected by mutations. We show non-genetic heterogeneity of the PDOs and upregulation of stemness and differentiation genes by kinase inhibitors. Using imaging mass-cytometry-based profiling of the primary tumors, we characterize the tumor microenvironment (TME) and determine spatial heterocellular crosstalk and tumor-immune cell interactions. Collectively, we provide a framework for inferring tumor cell intrinsic signaling and external signaling from the TME to inform precision (immuno-) oncology in CRC.
Keywords: Cancer; Cancer systems biology; Proteomics.
© 2023 The Author(s).
Conflict of interest statement
H.C. is an inventor on several patents related to organoid technology; his full disclosure is given at https://www.uu.nl/staff/JCClevers/. H.C. is currently head of pharma Research Early Development (pRED) at Roche. H.C. holds several patents on organoid technology. Their application numbers, followed by their publication numbers (if applicable), are as follows: PCT/NL2008/050543, WO2009/022907; PCT/NL2010/000017, WO2010/090513; PCT/IB2011/002167, WO2012/014076; PCT/IB2012/052950, WO2012/168930; PCT/EP2015/060815, WO2015/173425; PCT/EP2015/077990, WO2016/083613; PCT/EP2015/077988, WO2016/083612; PCT/EP2017/054797, WO2017/149025; PCT/EP2017/065101, WO2017/220586; PCT/EP2018/086716, n/a; and GB1819224.5, n/a.
Figures



References
-
- Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA. Cancer J. Clin. 2021;71:209–249. - PubMed
-
- Di Nicolantonio F., Vitiello P.P., Marsoni S., Siena S., Tabernero J., Trusolino L., Bernards R., Bardelli A. Precision oncology in metastatic colorectal cancer - from biology to medicine. Nat. Rev. Clin. Oncol. 2021;18:506–525. - PubMed
-
- Galon J., Costes A., Sanchez-Cabo F., Kirilovsky A., Mlecnik B., Lagorce-Pagès C., Tosolini M., Camus M., Berger A., Wind P., et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–1964. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

