INX-315, a Selective CDK2 Inhibitor, Induces Cell Cycle Arrest and Senescence in Solid Tumors
- PMID: 38047585
- PMCID: PMC10905675
- DOI: 10.1158/2159-8290.CD-23-0954
INX-315, a Selective CDK2 Inhibitor, Induces Cell Cycle Arrest and Senescence in Solid Tumors
Erratum in
-
Correction: INX-315, a Selective CDK2 Inhibitor, Induces Cell Cycle Arrest and Senescence in Solid Tumors.Cancer Discov. 2025 Oct 6;15(10):2185. doi: 10.1158/2159-8290.CD-25-1502. Cancer Discov. 2025. PMID: 41047844 Free PMC article. No abstract available.
Abstract
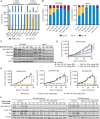
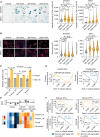
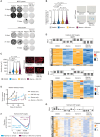
Cyclin-dependent kinase 2 (CDK2) is thought to play an important role in driving proliferation of certain cancers, including those harboring CCNE1 amplification and breast cancers that have acquired resistance to CDK4/6 inhibitors (CDK4/6i). The precise impact of pharmacologic inhibition of CDK2 is not known due to the lack of selective CDK2 inhibitors. Here we describe INX-315, a novel and potent CDK2 inhibitor with high selectivity over other CDK family members. Using cell-based assays, patient-derived xenografts (PDX), and transgenic mouse models, we show that INX-315 (i) promotes retinoblastoma protein hypophosphorylation and therapy-induced senescence (TIS) in CCNE1-amplified tumors, leading to durable control of tumor growth; (ii) overcomes breast cancer resistance to CDK4/6i, restoring cell cycle control while reinstating the chromatin architecture of CDK4/6i-induced TIS; and (iii) delays the onset of CDK4/6i resistance in breast cancer by driving deeper suppression of E2F targets. Our results support the clinical development of selective CDK2 inhibitors.
Significance: INX-315 is a novel, selective inhibitor of CDK2. Our preclinical studies demonstrate activity for INX-315 in both CCNE1-amplified cancers and CDK4/6i-resistant breast cancer. In each case, CDK2 inhibition induces cell cycle arrest and a phenotype resembling cellular senescence. Our data support the development of selective CDK2 inhibitors in clinical trials. See related commentary by Watts and Spencer, p. 386. This article is featured in Selected Articles from This Issue, p. 384.
©2023 The Authors; Published by the American Association for Cancer Research.
Figures



References
-
- Spring LM, Wander SA, Andre F, Moy B, Turner NC, Bardia A. Cyclin-dependent kinase 4 and 6 inhibitors for hormone receptor-positive breast cancer: past, present, and future. Lancet 2020;395:817–27. - PubMed
-
- Tadesse S, Anshabo AT, Portman N, Lim E, Tilley W, Caldon CE, et al. Targeting CDK2 in cancer: challenges and opportunities for therapy. Drug Discov Today 2020;25:406–13. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

