The role of N-glycosylation in spike antigenicity for the SARS-CoV-2 gamma variant
- PMID: 38048640
- PMCID: PMC10969516
- DOI: 10.1093/glycob/cwad097
The role of N-glycosylation in spike antigenicity for the SARS-CoV-2 gamma variant
Abstract
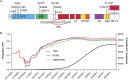
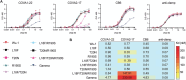
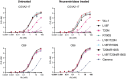
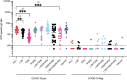
The emergence of SARS-CoV-2 variants alters the efficacy of existing immunity towards the viral spike protein, whether acquired from infection or vaccination. Mutations that impact N-glycosylation of spike may be particularly important in influencing antigenicity, but their consequences are difficult to predict. Here, we compare the glycosylation profiles and antigenicity of recombinant viral spike of ancestral Wu-1 and the Gamma strain, which has two additional N-glycosylation sites due to amino acid substitutions in the N-terminal domain (NTD). We found that a mutation at residue 20 from threonine to asparagine within the NTD caused the loss of NTD-specific antibody COVA2-17 binding. Glycan site-occupancy analyses revealed that the mutation resulted in N-glycosylation switching to the new sequon at N20 from the native N17 site. Site-specific glycosylation profiles demonstrated distinct glycoform differences between Wu-1, Gamma, and selected NTD variant spike proteins, but these did not affect antibody binding. Finally, we evaluated the specificity of spike proteins against convalescent COVID-19 sera and found reduced cross-reactivity against some mutants, but not Gamma spike compared to Wuhan spike. Our results illustrate the impact of viral divergence on spike glycosylation and SARS-CoV-2 antibody binding profiles.
Keywords: antibodies; coronavirus; glycoprotein; glycoproteomics; mass spectrometry.
© The Author(s) 2023. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures



References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

