PI3K signaling promotes formation of lipid-laden foamy macrophages at the spinal cord injury site
- PMID: 38049013
- PMCID: PMC10804283
- DOI: 10.1016/j.nbd.2023.106370
PI3K signaling promotes formation of lipid-laden foamy macrophages at the spinal cord injury site
Abstract
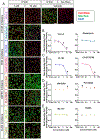
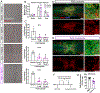
After spinal cord injury (SCI), infiltrating macrophages undergo excessive phagocytosis of myelin and cellular debris, forming lipid-laden foamy macrophages. To understand their role in the cellular pathology of SCI, investigation of the foamy macrophage phenotype in vitro revealed a pro-inflammatory profile, increased reactive oxygen species (ROS) production, and mitochondrial dysfunction. Bioinformatic analysis identified PI3K as a regulator of inflammation in foamy macrophages, and inhibition of this pathway decreased their lipid content, inflammatory cytokines, and ROS production. Macrophage-specific inhibition of PI3K using liposomes significantly decreased foamy macrophages at the injury site after a mid-thoracic contusive SCI in mice. RNA sequencing and in vitro analysis of foamy macrophages revealed increased autophagy and decreased phagocytosis after PI3K inhibition as potential mechanisms for reduced lipid accumulation. Together, our data suggest that the formation of pro-inflammatory foamy macrophages after SCI is due to the activation of PI3K signaling, which increases phagocytosis and decreases autophagy.
Keywords: Autophagy; Foamy macrophages; Lipids; Neuroinflammation; PI3K; Spinal cord injury.
Copyright © 2023. Published by Elsevier Inc.
Conflict of interest statement
Declaration of Competing Interest The authors declare no competing interests exist.
Figures







Similar articles
-
Myelin and non-myelin debris contribute to foamy macrophage formation after spinal cord injury.Neurobiol Dis. 2022 Feb;163:105608. doi: 10.1016/j.nbd.2021.105608. Epub 2021 Dec 31. Neurobiol Dis. 2022. PMID: 34979258 Free PMC article.
-
Cytosolic phospholipase A2 in infiltrating monocyte derived macrophages does not impair recovery after spinal cord injury in female mice.Sci Rep. 2025 Jan 2;15(1):1. doi: 10.1038/s41598-024-84936-6. Sci Rep. 2025. PMID: 39747330 Free PMC article.
-
Interleukin-3 Modulates Macrophage Phagocytic Activity and Promotes Spinal Cord Injury Repair.CNS Neurosci Ther. 2024 Dec;30(12):e70181. doi: 10.1111/cns.70181. CNS Neurosci Ther. 2024. PMID: 39697159 Free PMC article.
-
Spatial and temporal activation of spinal glial cells: role of gliopathy in central neuropathic pain following spinal cord injury in rats.Exp Neurol. 2012 Apr;234(2):362-72. doi: 10.1016/j.expneurol.2011.10.010. Epub 2011 Oct 21. Exp Neurol. 2012. PMID: 22036747 Free PMC article.
-
The Role of Tumor Necrosis Factor Following Spinal Cord Injury: A Systematic Review.Cell Mol Neurobiol. 2023 Apr;43(3):925-950. doi: 10.1007/s10571-022-01229-0. Epub 2022 May 23. Cell Mol Neurobiol. 2023. PMID: 35604578 Free PMC article.
Cited by
-
Molecular pathology of acute spinal cord injury in middle-aged mice.bioRxiv [Preprint]. 2025 May 14:2025.05.08.652873. doi: 10.1101/2025.05.08.652873. bioRxiv. 2025. Update in: J Neuroinflammation. 2025 Jul 13;22(1):181. doi: 10.1186/s12974-025-03494-4. PMID: 40463189 Free PMC article. Updated. Preprint.
-
Biomaterials targeting the microenvironment for spinal cord injury repair: progression and perspectives.Front Cell Neurosci. 2024 May 9;18:1362494. doi: 10.3389/fncel.2024.1362494. eCollection 2024. Front Cell Neurosci. 2024. PMID: 38784712 Free PMC article. Review.
-
The protective PLCγ2-P522R variant mitigates Alzheimer's disease-associated pathologies by enhancing beneficial microglial functions.J Neuroinflammation. 2025 Mar 5;22(1):64. doi: 10.1186/s12974-025-03387-6. J Neuroinflammation. 2025. PMID: 40038760 Free PMC article.
-
Molecular pathology of acute spinal cord injury in middle-aged mice.J Neuroinflammation. 2025 Jul 13;22(1):181. doi: 10.1186/s12974-025-03494-4. J Neuroinflammation. 2025. PMID: 40653490 Free PMC article.
-
Nitroxidative Stress, Cell-Signaling Pathways, and Manganese Porphyrins: Therapeutic Potential in Neuropathic Pain.Int J Mol Sci. 2025 Feb 26;26(5):2050. doi: 10.3390/ijms26052050. Int J Mol Sci. 2025. PMID: 40076672 Free PMC article. Review.
References
-
- Aderem A, Underhill DM, 1999. Mechanisms of phagocytosis in macrophages. Annu. Rev. Immunol 17, 593–623. - PubMed
-
- Alexa A, Rahnenfuhrer J, 2023. topGO: Enrichment Analysis for Gene Ontology. doi: 10.18129/B9.bioc.topGO. R package version 2.54.0, https://bioconductor.org/packages/topGO. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

