A structure of the relict phycobilisome from a thylakoid-free cyanobacterium
- PMID: 38049400
- PMCID: PMC10696076
- DOI: 10.1038/s41467-023-43646-9
A structure of the relict phycobilisome from a thylakoid-free cyanobacterium
Erratum in
-
Author Correction: A structure of the relict phycobilisome from a thylakoid-free cyanobacterium.Nat Commun. 2023 Dec 19;14(1):8436. doi: 10.1038/s41467-023-44394-6. Nat Commun. 2023. PMID: 38114511 Free PMC article. No abstract available.
Abstract
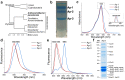
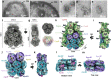
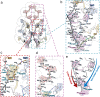
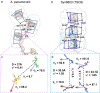
Phycobilisomes (PBS) are antenna megacomplexes that transfer energy to photosystems II and I in thylakoids. PBS likely evolved from a basic, inefficient form into the predominant hemidiscoidal shape with radiating peripheral rods. However, it has been challenging to test this hypothesis because ancestral species are generally inaccessible. Here we use spectroscopy and cryo-electron microscopy to reveal a structure of a "paddle-shaped" PBS from a thylakoid-free cyanobacterium that likely retains ancestral traits. This PBS lacks rods and specialized ApcD and ApcF subunits, indicating relict characteristics. Other features include linkers connecting two chains of five phycocyanin hexamers (CpcN) and two core subdomains (ApcH), resulting in a paddle-shaped configuration. Energy transfer calculations demonstrate that chains are less efficient than rods. These features may nevertheless have increased light absorption by elongating PBS before multilayered thylakoids with hemidiscoidal PBS evolved. Our results provide insights into the evolution and diversification of light-harvesting strategies before the origin of thylakoids.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Light-induced structural adaptation of the bundle-shaped phycobilisome from thylakoid-lacking cyanobacterium Gloeobacter violaceus.Nat Commun. 2025 Jul 1;16(1):5956. doi: 10.1038/s41467-025-60673-w. Nat Commun. 2025. PMID: 40593595 Free PMC article.
-
The membrane-associated CpcG2-phycobilisome in Synechocystis: a new photosystem I antenna.Plant Physiol. 2007 Jun;144(2):1200-10. doi: 10.1104/pp.107.099267. Epub 2007 Apr 27. Plant Physiol. 2007. PMID: 17468217 Free PMC article.
-
Significant energy transfer from CpcG2-phycobilisomes to photosystem I in the cyanobacterium Synechococcus sp. PCC 7002 in the absence of ApcD-dependent state transitions.FEBS Lett. 2012 Jul 30;586(16):2342-5. doi: 10.1016/j.febslet.2012.05.038. Epub 2012 May 31. FEBS Lett. 2012. PMID: 22659186
-
The amazing phycobilisome.Biochim Biophys Acta Bioenerg. 2020 Apr 1;1861(4):148047. doi: 10.1016/j.bbabio.2019.07.002. Epub 2019 Jul 12. Biochim Biophys Acta Bioenerg. 2020. PMID: 31306623 Review.
-
Phycobilisomes and Phycobiliproteins in the Pigment Apparatus of Oxygenic Photosynthetics: From Cyanobacteria to Tertiary Endosymbiosis.Int J Mol Sci. 2023 Jan 24;24(3):2290. doi: 10.3390/ijms24032290. Int J Mol Sci. 2023. PMID: 36768613 Free PMC article. Review.
Cited by
-
The structural basis for light harvesting in organisms producing phycobiliproteins.Plant Cell. 2024 Oct 3;36(10):4036-4064. doi: 10.1093/plcell/koae126. Plant Cell. 2024. PMID: 38652697 Free PMC article. Review.
-
N-Terminal domain homologs of the orange carotenoid protein increase quenching of cyanobacterial phycobilisomes.Plant Physiol. 2025 Apr 30;198(1):kiae531. doi: 10.1093/plphys/kiae531. Plant Physiol. 2025. PMID: 39365917 Free PMC article.
-
Thylakoids for enhanced photodynamic therapy in hypoxic tumours.Sci Technol Adv Mater. 2025 Jul 25;26(1):2537000. doi: 10.1080/14686996.2025.2537000. eCollection 2025. Sci Technol Adv Mater. 2025. PMID: 40786231 Free PMC article. Review.
-
Archaean green-light environments drove the evolution of cyanobacteria's light-harvesting system.Nat Ecol Evol. 2025 Apr;9(4):599-612. doi: 10.1038/s41559-025-02637-3. Epub 2025 Feb 18. Nat Ecol Evol. 2025. PMID: 39966498 Free PMC article.
-
Light-induced structural adaptation of the bundle-shaped phycobilisome from thylakoid-lacking cyanobacterium Gloeobacter violaceus.Nat Commun. 2025 Jul 1;16(1):5956. doi: 10.1038/s41467-025-60673-w. Nat Commun. 2025. PMID: 40593595 Free PMC article.
References
-
- Overmann, J. & Garcia-Pichel, F. in The Prokaryotes: Prokaryotic Communities and Ecophysiology (eds E. Rosenberg et al.) 203–257 (Springer Berlin Heidelberg, 2013).
-
- Sidler, W. A. in The Molecular Biology of Cyanobacteria In: Advances in Photosynthesis, Vol. 1 (ed D. A. Bryant) 139–216 (Springer Netherlands, 1994).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

