Making sense of chemical space network shows signs of criticality
- PMID: 38049451
- PMCID: PMC10696027
- DOI: 10.1038/s41598-023-48107-3
Making sense of chemical space network shows signs of criticality
Abstract
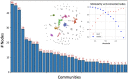
Chemical space modelling has great importance in unveiling and visualising latent information, which is critical in predictive toxicology related to drug discovery process. While the use of traditional molecular descriptors and fingerprints may suffer from the so-called curse of dimensionality, complex networks are devoid of the typical drawbacks of coordinate-based representations. Herein, we use chemical space networks (CSNs) to analyse the case of the developmental toxicity (Dev Tox), which remains a challenging endpoint for the difficulty of gathering enough reliable data despite very important for the protection of the maternal and child health. Our study proved that the Dev Tox CSN has a complex non-random organisation and can thus provide a wealth of meaningful information also for predictive purposes. At a phase transition, chemical similarities highlight well-established toxicophores, such as aryl derivatives, mostly neurotoxic hydantoins, barbiturates and amino alcohols, steroids, and volatile organic compounds ether-like chemicals, which are strongly suspected of the Dev Tox onset and can thus be employed as effective alerts for prioritising chemicals before testing.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
LinkOut - more resources
Full Text Sources

