Characterisation of FLT3 alterations in childhood acute lymphoblastic leukaemia
- PMID: 38049555
- PMCID: PMC10803556
- DOI: 10.1038/s41416-023-02511-8
Characterisation of FLT3 alterations in childhood acute lymphoblastic leukaemia
Abstract
Background: Alterations of FLT3 are among the most common driver events in acute leukaemia with important clinical implications, since it allows patient classification into prognostic groups and the possibility of personalising therapy thanks to the availability of FLT3 inhibitors. Most of the knowledge on FLT3 implications comes from the study of acute myeloid leukaemia and so far, few studies have been performed in other leukaemias.
Methods: A comprehensive genomic (DNA-seq in 267 patients) and transcriptomic (RNA-seq in 160 patients) analysis of FLT3 in 342 childhood acute lymphoblastic leukaemia (ALL) patients was performed. Mutations were functionally characterised by in vitro experiments.
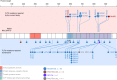
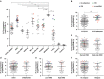
Results: Point mutations (PM) and internal tandem duplications (ITD) were detected in 4.3% and 2.7% of the patients, respectively. A new activating mutation of the TKD, G846D, conferred oncogenic properties and sorafenib resistance. Moreover, a novel alteration involving the circularisation of read-through transcripts (rt-circRNAs) was observed in 10% of the cases. Patients presenting FLT3 alterations exhibited higher levels of the receptor. In addition, patients with ZNF384- and MLL/KMT2A-rearranged ALL, as well as hyperdiploid subtype, overexpressed FLT3.
Discussion: Our results suggest that specific ALL subgroups may also benefit from a deeper understanding of the biology of FLT3 alterations and their clinical implications.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

