Rational optimization of a transcription factor activation domain inhibitor
- PMID: 38049566
- PMCID: PMC10716049
- DOI: 10.1038/s41594-023-01159-5
Rational optimization of a transcription factor activation domain inhibitor
Abstract
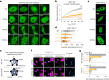
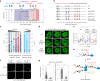
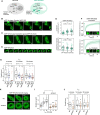
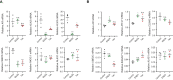
Transcription factors are among the most attractive therapeutic targets but are considered largely 'undruggable' in part due to the intrinsically disordered nature of their activation domains. Here we show that the aromatic character of the activation domain of the androgen receptor, a therapeutic target for castration-resistant prostate cancer, is key for its activity as transcription factor, allowing it to translocate to the nucleus and partition into transcriptional condensates upon activation by androgens. On the basis of our understanding of the interactions stabilizing such condensates and of the structure that the domain adopts upon condensation, we optimized the structure of a small-molecule inhibitor previously identified by phenotypic screening. The optimized compounds had more affinity for their target, inhibited androgen-receptor-dependent transcriptional programs, and had an antitumorigenic effect in models of castration-resistant prostate cancer in cells and in vivo. These results suggest that it is possible to rationally optimize, and potentially even to design, small molecules that target the activation domains of oncogenic transcription factors.
© 2023. The Author(s).
Conflict of interest statement
M.F.-V. is an employee of Dewpoint Therapeutics. M.P., M.L., L.Z., H.D. and M.B. are employees of Nuage Therapeutics. M.B., D.H. and X.S. are founders of Nuage Therapeutics. D.H. and X.S. are scientific advisors of Nuage Therapeutics. The remaining authors declare no competing interests.
Figures










References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

