SNORA56-mediated pseudouridylation of 28 S rRNA inhibits ferroptosis and promotes colorectal cancer proliferation by enhancing GCLC translation
- PMID: 38049865
- PMCID: PMC10696674
- DOI: 10.1186/s13046-023-02906-8
SNORA56-mediated pseudouridylation of 28 S rRNA inhibits ferroptosis and promotes colorectal cancer proliferation by enhancing GCLC translation
Abstract
Background: Colorectal cancer (CRC) is one of the most common malignancies and is characterized by reprogrammed metabolism. Ferroptosis, a programmed cell death dependent on iron, has emerged as a promising strategy for CRC treatment. Although small nucleolar RNAs are extensively involved in carcinogenesis, it is unclear if they regulate ferroptosis during CRC pathogenesis.
Methods: The dysregulated snoRNAs were identified using published sequencing data of CRC tissues. The expression of the candidate snoRNAs, host gene and target gene were assessed by real-time quantitative PCR (RT-qPCR), fluorescence in situ hybridization (FISH), immunohistochemistry (IHC) and western blots. The biological function of critical molecules was investigated using in vitro and in vivo strategies including Cell Counting Kit-8 (CCK8), colony formation assay, flow cytometry, Fe2+/Fe3+, GSH/GSSG and the xenograft mice models. The ribosomal activities were determined by polysome profiling and O-propargyl-puromycin (OP-Puro) assay. The proteomics was conducted to clarify the downstream targets and the underlying mechanisms were validated by IHC, Pearson correlation analysis, protein stability and rescue assays. The clinical significance of the snoRNA was explored using the Cox proportional hazard model, receiver operating characteristic (ROC) and survival analysis.
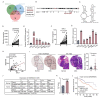
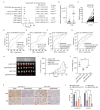
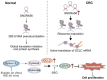
Results: Here, we investigated the SNORA56, which was elevated in CRC tissues and plasma, and correlated with CRC prognosis. SNORA56 deficiency in CRC impaired proliferation and triggered ferroptosis, resulting in reduced tumorigenesis. Mechanistically, SNORA56 mediated the pseudouridylation of 28 S rRNA at the U1664 site and promoted the translation of the catalytic subunit of glutamate cysteine ligase (GCLC), an indispensable rate-limiting enzyme in the biosynthesis of glutathione, which can inhibit ferroptosis by suppressing lipid peroxidation.
Conclusions: Therefore, the SNORA56/28S rRNA/GCLC axis stimulates CRC progression by inhibiting the accumulation of cellular peroxides, and it may provide biomarker and therapeutic applications in CRC.
Keywords: Biomarker; Colorectal cancer; Ferroptosis; GCLC; Proliferation; Pseudouridylation; SNORA56; Therapy.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- Patel SG, Karlitz JJ, Yen T, Lieu CH, Boland CR. The rising tide of early-onset Colorectal cancer: a comprehensive review of epidemiology, clinical features, biology, risk factors, prevention, and early detection. Lancet Gastroenterol Hepatol. 2022;7:262–74. doi: 10.1016/S2468-1253(21)00426-X. - DOI - PubMed
MeSH terms
Substances
Grants and funding
- 81930066/the National Natural Science Foundation of China
- 82172357/the National Natural Science Foundation of China
- 82293662/the National Natural Science Foundation of China
- 2019CXJQ03/the Innovation Group Project of Shanghai Municipal Health Commission
- 22JC1402300/Science and Technology Innovation Plan Of Shanghai Science and Technology Commission
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

