Hypoxia-inducible microRNA-155 negatively regulates epithelial barrier in eosinophilic esophagitis by suppressing tight junction claudin-7
- PMID: 38050671
- PMCID: PMC10699209
- DOI: 10.1096/fj.202301934R
Hypoxia-inducible microRNA-155 negatively regulates epithelial barrier in eosinophilic esophagitis by suppressing tight junction claudin-7
Abstract
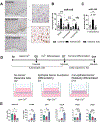
MicroRNA (miRNA)-mediated mRNA regulation directs many homeostatic and pathological processes, but how miRNAs coordinate aberrant esophageal inflammation during eosinophilic esophagitis (EoE) is poorly understood. Here, we report a deregulatory axis where microRNA-155 (miR-155) regulates epithelial barrier dysfunction by selectively constraining tight junction CLDN7 (claudin-7). MiR-155 is elevated in the esophageal epithelium of biopsies from patients with active EoE and in cell culture models. MiR-155 localization using in situ hybridization (ISH) in patient biopsies and intra-epithelial compartmentalization of miR-155 show expression predominantly within the basal epithelia. Epithelial miR-155 activity was evident through diminished target gene expression in 3D organotypic cultures, particularly in relatively undifferentiated basal cell states. Mechanistically, generation of a novel cell line with enhanced epithelial miR-155 stable overexpression induced a functionally deficient epithelial barrier in 3D air-liquid interface epithelial cultures measured by transepithelial electrical resistance (TEER). Histological assessment of 3D esophageal organoid cultures overexpressing miR-155 showed notable dilated intra-epithelial spaces. Unbiased RNA-sequencing analysis and immunofluorescence determined a defect in epithelial barrier tight junctions and revealed a selective reduction in the expression of critical esophageal tight junction molecule, claudin-7. Together, our data reveal a previously unappreciated role for miR-155 in mediating epithelial barrier dysfunction in esophageal inflammation.
Keywords: Barrier; claudin-7; eosinophilic esophagitis; epithelium; hypoxia; miR-155; microRNA 155; tight junction.
© 2023 The Authors. The FASEB Journal published by Wiley Periodicals LLC on behalf of Federation of American Societies for Experimental Biology.
Conflict of interest statement
Conflict of Interest Statement
Authors have declared that no conflict of interest exists.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

