Circ-0000953 deficiency exacerbates podocyte injury and autophagy disorder by targeting Mir665-3p-Atg4b in diabetic nephropathy
- PMID: 38050963
- PMCID: PMC11135827
- DOI: 10.1080/15548627.2023.2286128
Circ-0000953 deficiency exacerbates podocyte injury and autophagy disorder by targeting Mir665-3p-Atg4b in diabetic nephropathy
Abstract
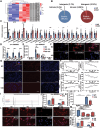
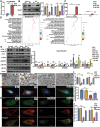
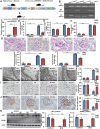
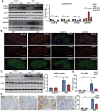
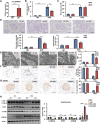
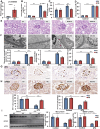
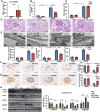
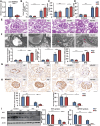
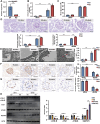
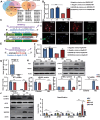
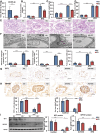
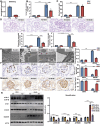
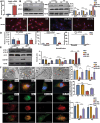
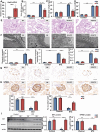
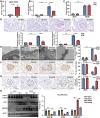
Circular RNAs (circRNAs) are special non-coding RNA (ncRNA) molecules that play a significant role in many diseases. However, the biogenesis and regulation of circRNAs in diabetic nephropathy (DN) are largely unknown. Here, we investigated the expression profile of circRNAs in kidney of DN mice through circular RNA sequencing (circRNA-seq). The renal biopsy samples of patients with DN had low circ -0,000,953 expression, which was significantly associated with renal function. Furthermore, loss-of-function and gain-of-function experiments were carried out to prove the role of circ -0,000,953 in DN. Podocyte conditional knockin (cKI) or systemic overexpression of circ -0,000,953 alleviated albuminuria and restored macroautophagy/autophagy in kidney of diabetic mice. However, circ -0,000,953 knockdown exacerbated albuminuria and podocyte injury. Mechanistically, we found circ -0,000,953 directly binds to Mir665-3p-Atg4b to perform its function. Silencing of Mir665-3p or overexpression of Atg4b recovered podocyte autophagy both in vitro and in vivo. To examine the cause of circ -0,000,953 downregulation in DN, bioinformatics prediction found that circ -0,000,953 sequence has a high possibility of containing an m6A methylation site. Additionally, METTL3 was proved to regulate the expression and methylation level of circ -0,000,953 through YTHDF2 (YTH N6-methyladenosine RNA binding protein 2). In conclusion, this study revealed that circ -0,000,953 regulates podocyte autophagy by targeting Mir665-3p-Atg4b in DN. Therefore, circ -0,000,953 is a potential biomarker for prevention and cure of DN.Abbreviation: CCL2/MCP-1: C-C motif chemokine ligand 2; ceRNA: competing endogenous RNA; circRNA: circular RNA; cKI: conditional knockin; cKO: conditional knockout; CRE: creatinine; DM: diabetes mellitus; DN: diabetic nephropathy; ESRD: end-stage renal disease; HG: high glucose; IF: immunofluorescence; MAP1LC3/LC3B: microtubule-associated protein 1 light chain 3 beta; MPC5: mouse podocyte clone 5; MTECs: mouse tubular epithelial cells; MTOR: mechanistic target of rapamycin kinase; NC: normal control; ncRNA: non-coding RNA; NPHS1: nephrosis 1, nephrin; NPHS2: nephrosis 2, podocin; PAS: periodic acid-Schiff; RELA/p65: v-rel reticuloendotheliosis viral oncogene homolog A (avian); SDs: slit diaphragm proteins; Seq: sequencing; STZ: streptozotocin; SV40: SV40-MES13-cells, mouse mesangial cell line; T1D: type 1 diabetes mellitus; T2D: type 2 diabetes mellitus; TEM: transmission electron microscopy; TNF/TNF-α: tumor necrosis factor; VECs: vascular endothelial cells; WT1: WT1 transcription factor; YTHDF2: YTH N6-methyladenosine RNA binding protein 2.
Keywords: Atg4b; autophagy; circRNA; diabetic nephropathy; m6A; podocytes.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures
Similar articles
-
miR-188-3p abolishes germacrone-mediated podocyte protection in a mouse model of diabetic nephropathy in type I diabetes through triggering mitochondrial injury.Bioengineered. 2022 Jan;13(1):774-788. doi: 10.1080/21655979.2021.2012919. Bioengineered. 2022. Retraction in: Bioengineered. 2024 Dec;15(1):2299558. doi: 10.1080/21655979.2024.2299558. PMID: 34847832 Free PMC article. Retracted.
-
Circ_0000285 promotes podocyte injury through sponging miR-654-3p and activating MAPK6 in diabetic nephropathy.Gene. 2020 Jul 15;747:144661. doi: 10.1016/j.gene.2020.144661. Epub 2020 Apr 7. Gene. 2020. PMID: 32275999
-
METTL3 promotes podocyte pyroptosis in diabetic nephropathy through N6-methyladenosine modification of TRIM29 mRNA.Ren Fail. 2025 Dec;47(1):2497492. doi: 10.1080/0886022X.2025.2497492. Epub 2025 May 5. Ren Fail. 2025. PMID: 40325507 Free PMC article.
-
The role and mechanism of m6A methylation in diabetic nephropathy.Life Sci. 2025 Feb 15;363:123355. doi: 10.1016/j.lfs.2024.123355. Epub 2025 Jan 6. Life Sci. 2025. PMID: 39778764 Review.
-
Crosstalk between non-coding RNA and apoptotic signaling in diabetic nephropathy.Biochem Pharmacol. 2024 Dec;230(Pt 3):116621. doi: 10.1016/j.bcp.2024.116621. Epub 2024 Nov 13. Biochem Pharmacol. 2024. PMID: 39542182 Review.
Cited by
-
N6-methyladenine RNA methylation epigenetic modification and diabetic microvascular complications.Front Endocrinol (Lausanne). 2024 Sep 4;15:1462146. doi: 10.3389/fendo.2024.1462146. eCollection 2024. Front Endocrinol (Lausanne). 2024. PMID: 39296713 Free PMC article. Review.
-
Decoding m6A RNA methylation in kidney disorders: from molecular insights to therapeutic strategies.J Transl Med. 2025 Jul 10;23(1):771. doi: 10.1186/s12967-025-06817-4. J Transl Med. 2025. PMID: 40640897 Free PMC article. Review.
-
Circular RNA APP contributes to Alzheimer's disease pathogenesis by modulating microglial polarization via miR-1906/CLIC1 axis.Alzheimers Res Ther. 2025 Feb 14;17(1):44. doi: 10.1186/s13195-025-01698-7. Alzheimers Res Ther. 2025. PMID: 39953602 Free PMC article.
-
Understanding the Roles of Non-coding RNAs and Exosomal Non-Coding RNAs in Diabetic Nephropathy.Curr Mol Med. 2025;25(5):537-555. doi: 10.2174/0115665240287631240321072504. Curr Mol Med. 2025. PMID: 38591211 Review.
-
circ0066187 promotes pulmonary fibrogenesis through targeting STAT3-mediated metabolism signal pathway.Cell Mol Life Sci. 2025 Feb 19;82(1):79. doi: 10.1007/s00018-025-05613-z. Cell Mol Life Sci. 2025. PMID: 39969586 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
