Fission Yeast TORC1 Promotes Cell Proliferation through Sfp1, a Transcription Factor Involved in Ribosome Biogenesis
- PMID: 38051102
- PMCID: PMC10761059
- DOI: 10.1080/10985549.2023.2282349
Fission Yeast TORC1 Promotes Cell Proliferation through Sfp1, a Transcription Factor Involved in Ribosome Biogenesis
Abstract
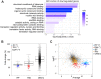
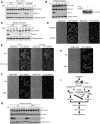
Target of rapamycin complex 1 (TORC1) is activated in response to nutrient availability and growth factors, promoting cellular anabolism and proliferation. To explore the mechanism of TORC1-mediated proliferation control, we performed a genetic screen in fission yeast and identified Sfp1, a zinc-finger transcription factor, as a multicopy suppressor of temperature-sensitive TORC1 mutants. Our observations suggest that TORC1 phosphorylates Sfp1 and protects Sfp1 from proteasomal degradation. Transcription analysis revealed that Sfp1 positively regulates genes involved in ribosome production together with two additional transcription factors, Ifh1/Crf1 and Fhl1. Ifh1 physically interacts with Fhl1, and the nuclear localization of Ifh1 is regulated in response to nutrient levels in a manner dependent on TORC1 and Sfp1. Taken together, our data suggest that the transcriptional regulation of the genes involved in ribosome biosynthesis by Sfp1, Ifh1, and Fhl1 is one of the key pathways through which nutrient-activated TORC1 promotes cell proliferation.
Keywords: Sfp1; TORC1; fission yeast; rapamycin; ribosome; transcription factor.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
