An mTurq2-Col4a1 mouse model allows for live visualization of mammalian basement membrane development
- PMID: 38051393
- PMCID: PMC10697824
- DOI: 10.1083/jcb.202309074
An mTurq2-Col4a1 mouse model allows for live visualization of mammalian basement membrane development
Abstract
Basement membranes (BMs) are specialized sheets of extracellular matrix that underlie epithelial and endothelial tissues. BMs regulate the traffic of cells and molecules between compartments, and participate in signaling, cell migration, and organogenesis. The dynamics of mammalian BMs, however, are poorly understood, largely due to a lack of models in which core BM components are endogenously labeled. Here, we describe the mTurquoise2-Col4a1 mouse in which we fluorescently tag collagen IV, the main component of BMs. Using an innovative planar-sagittal live imaging technique to visualize the BM of developing skin, we directly observe BM deformation during hair follicle budding and basal progenitor cell divisions. The BM's inherent pliability enables dividing cells to remain attached to and deform the BM, rather than lose adhesion as generally thought. Using FRAP, we show BM collagen IV is extremely stable, even during periods of rapid epidermal growth. These findings demonstrate the utility of the mTurq2-Col4a1 mouse to shed new light on mammalian BM developmental dynamics.
© 2023 Jones et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures
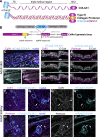


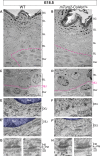




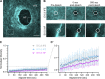
Update of
-
A Window into Mammalian Basement Membrane Development: Insights from the mTurq2-Col4a1 Mouse Model.bioRxiv [Preprint]. 2023 Sep 27:2023.09.27.559396. doi: 10.1101/2023.09.27.559396. bioRxiv. 2023. Update in: J Cell Biol. 2024 Feb 5;223(2):e202309074. doi: 10.1083/jcb.202309074. PMID: 37808687 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

