Commensal Oral Microbiota, Disease Severity, and Mortality in Fibrotic Lung Disease
- PMID: 38051927
- PMCID: PMC11092942
- DOI: 10.1164/rccm.202308-1357OC
Commensal Oral Microbiota, Disease Severity, and Mortality in Fibrotic Lung Disease
Abstract
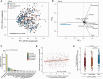
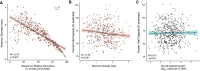
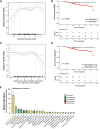
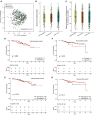
Rationale: Oral microbiota associate with diseases of the mouth and serve as a source of lung microbiota. However, the role of oral microbiota in lung disease is unknown. Objectives: To determine associations between oral microbiota and disease severity and death in idiopathic pulmonary fibrosis (IPF). Methods: We analyzed 16S rRNA gene and shotgun metagenomic sequencing data of buccal swabs from 511 patients with IPF in the multicenter CleanUP-IPF (Study of Clinical Efficacy of Antimicrobial Therapy Strategy Using Pragmatic Design in IPF) trial. Buccal swabs were collected from usual care and antimicrobial cohorts. Microbiome data were correlated with measures of disease severity using principal component analysis and linear regression models. Associations between the buccal microbiome and mortality were determined using Cox additive models, Kaplan-Meier analysis, and Cox proportional hazards models. Measurements and Main Results: Greater buccal microbial diversity associated with lower FVC at baseline (mean difference, -3.60; 95% confidence interval [CI], -5.92 to -1.29% predicted FVC per 1-unit increment). The buccal proportion of Streptococcus correlated positively with FVC (mean difference, 0.80; 95% CI, 0.16 to 1.43% predicted per 10% increase) (n = 490). Greater microbial diversity was associated with an increased risk of death (hazard ratio, 1.73; 95% CI, 1.03-2.90), whereas a greater proportion of Streptococcus was associated with a reduced risk of death (HR, 0.85; 95% CI, 0.73 to 0.99). The Streptococcus genus was mainly composed of Streptococcus mitis species. Conclusions: Increasing buccal microbial diversity predicts disease severity and death in IPF. The oral commensal S. mitis spp associates with preserved lung function and improved survival.
Keywords: buccal microbiome; disease progression; idiopathic pulmonary fibrosis.
Figures
Comment in
-
Looking Beyond the Lower Airways for Microbes Affecting Pulmonary Fibrosis.Am J Respir Crit Care Med. 2024 May 1;209(9):1054-1056. doi: 10.1164/rccm.202312-2255ED. Am J Respir Crit Care Med. 2024. PMID: 38227734 Free PMC article. No abstract available.
References
-
- Yan Z, Chen B, Yang Y, Yi X, Wei M, Ecklu-Mensah G, et al. Multi-omics analyses of airway host-microbe interactions in chronic obstructive pulmonary disease identify potential therapeutic interventions. Nat Microbiol . 2022;7:1361–1375. - PubMed
-
- Lederer DJ, Martinez FJ. Idiopathic pulmonary fibrosis. N Engl J Med . 2018;379:797–798. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

