Genome-wide association study meta-analysis of dizygotic twinning illuminates genetic regulation of female fecundity
- PMID: 38052102
- PMCID: PMC10767824
- DOI: 10.1093/humrep/dead247
Genome-wide association study meta-analysis of dizygotic twinning illuminates genetic regulation of female fecundity
Abstract
Study question: Which genetic factors regulate female propensity for giving birth to spontaneous dizygotic (DZ) twins?
Summary answer: We identified four new loci, GNRH1, FSHR, ZFPM1, and IPO8, in addition to previously identified loci, FSHB and SMAD3.
What is known already: The propensity to give birth to DZ twins runs in families. Earlier, we reported that FSHB and SMAD3 as associated with DZ twinning and female fertility measures.
Study design, size, duration: We conducted a genome-wide association meta-analysis (GWAMA) of mothers of spontaneous dizygotic (DZ) twins (8265 cases, 264 567 controls) and of independent DZ twin offspring (26 252 cases, 417 433 controls).
Participants/materials, setting, methods: Over 700 000 mothers of DZ twins, twin individuals and singletons from large cohorts in Australia/New Zealand, Europe, and the USA were carefully screened to exclude twins born after use of ARTs. Genetic association analyses by cohort were followed by meta-analysis, phenome wide association studies (PheWAS), in silico and in vivo annotations, and Zebrafish functional validation.
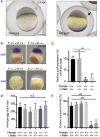
Main results and the role of chance: This study enlarges the sample size considerably from previous efforts, finding four genome-wide significant loci, including two novel signals and a further two novel genes that are implicated by gene level enrichment analyses. The novel loci, GNRH1 and FSHR, have well-established roles in female reproduction whereas ZFPM1 and IPO8 have not previously been implicated in female fertility. We found significant genetic correlations with multiple aspects of female reproduction and body size as well as evidence for significant selection against DZ twinning during human evolution. The 26 top single nucleotide polymorphisms (SNPs) from our GWAMA in European-origin participants weakly predicted the crude twinning rates in 47 non-European populations (r = 0.23 between risk score and population prevalence, s.e. 0.11, 1-tail P = 0.058) indicating that genome-wide association studies (GWAS) are needed in African and Asian populations to explore the causes of their respectively high and low DZ twinning rates. In vivo functional tests in zebrafish for IPO8 validated its essential role in female, but not male, fertility. In most regions, risk SNPs linked to known expression quantitative trait loci (eQTLs). Top SNPs were associated with in vivo reproductive hormone levels with the top pathways including hormone ligand binding receptors and the ovulation cycle.
Large scale data: The full DZT GWAS summary statistics will made available after publication through the GWAS catalog (https://www.ebi.ac.uk/gwas/).
Limitations, reasons for caution: Our study only included European ancestry cohorts. Inclusion of data from Africa (with the highest twining rate) and Asia (with the lowest rate) would illuminate further the biology of twinning and female fertility.
Wider implications of the findings: About one in 40 babies born in the world is a twin and there is much speculation on why twinning runs in families. We hope our results will inform investigations of ovarian response in new and existing ARTs and the causes of female infertility.
Study funding/competing interest(s): Support for the Netherlands Twin Register came from the Netherlands Organization for Scientific Research (NWO) and The Netherlands Organization for Health Research and Development (ZonMW) grants, 904-61-193, 480-04-004, 400-05-717, Addiction-31160008, 911-09-032, Biobanking and Biomolecular Resources Research Infrastructure (BBMRI.NL, 184.021.007), Royal Netherlands Academy of Science Professor Award (PAH/6635) to DIB, European Research Council (ERC-230374), Rutgers University Cell and DNA Repository (NIMH U24 MH068457-06), the Avera Institute, Sioux Falls, South Dakota (USA) and the National Institutes of Health (NIH R01 HD042157-01A1) and the Genetic Association Information Network (GAIN) of the Foundation for the National Institutes of Health and Grand Opportunity grants 1RC2 MH089951. The QIMR Berghofer Medical Research Institute (QIMR) study was supported by grants from the National Health and Medical Research Council (NHMRC) of Australia (241944, 339462, 389927, 389875, 389891, 389892, 389938, 443036, 442915, 442981, 496610, 496739, 552485, 552498, 1050208, 1075175). L.Y. is funded by Australian Research Council (Grant number DE200100425). The Minnesota Center for Twin and Family Research (MCTFR) was supported in part by USPHS Grants from the National Institute on Alcohol Abuse and Alcoholism (AA09367 and AA11886) and the National Institute on Drug Abuse (DA05147, DA13240, and DA024417). The Women's Genome Health Study (WGHS) was funded by the National Heart, Lung, and Blood Institute (HL043851 and HL080467) and the National Cancer Institute (CA047988 and UM1CA182913), with support for genotyping provided by Amgen. Data collection in the Finnish Twin Registry has been supported by the Wellcome Trust Sanger Institute, the Broad Institute, ENGAGE-European Network for Genetic and Genomic Epidemiology, FP7-HEALTH-F4-2007, grant agreement number 201413, National Institute of Alcohol Abuse and Alcoholism (grants AA-12502, AA-00145, AA-09203, AA15416, and K02AA018755) and the Academy of Finland (grants 100499, 205585, 118555, 141054, 264146, 308248, 312073 and 336823 to J. Kaprio). TwinsUK is funded by the Wellcome Trust, Medical Research Council, Versus Arthritis, European Union Horizon 2020, Chronic Disease Research Foundation (CDRF), Zoe Ltd and the National Institute for Health Research (NIHR) Clinical Research Network (CRN) and Biomedical Research Centre based at Guy's and St Thomas' NHS Foundation Trust in partnership with King's College London. For NESDA, funding was obtained from the Netherlands Organization for Scientific Research (Geestkracht program grant 10000-1002), the Center for Medical Systems Biology (CSMB, NVVO Genomics), Biobanking and Biomolecular Resources Research Infrastructure (BBMRI-NL), VU University's Institutes for Health and Care Research (EMGO+) and Neuroscience Campus Amsterdam, University Medical Center Groningen, Leiden University Medical Center, National Institutes of Health (NIH, ROI D0042157-01A, MH081802, Grand Opportunity grants 1 RC2 Ml-1089951 and IRC2 MH089995). Part of the genotyping and analyses were funded by the Genetic Association Information Network (GAIN) of the Foundation for the National Institutes of Health. Computing was supported by BiG Grid, the Dutch e-Science Grid, which is financially supported by NWO. Work in the Del Bene lab was supported by the Programme Investissements d'Avenir IHU FOReSIGHT (ANR-18-IAHU-01). C.R. was supported by an EU Horizon 2020 Marie Skłodowska-Curie Action fellowship (H2020-MSCA-IF-2014 #661527). H.S. and K.S. are employees of deCODE Genetics/Amgen. The other authors declare no competing financial interests.
Trial registration number: N/A.
Keywords: dizygotic twinning; female fecundity; fertility; genetics; genome-wide association analysis.
© The Author(s) 2023. Published by Oxford University Press on behalf of European Society of Human Reproduction and Embryology.
Conflict of interest statement
H.S. and K.S. are employees of deCODE Genetics/Amgen. A.C. is an employee of the Regeneron Genetics Center (Regeneron Pharmaceuticals) and may own stock to stock options. The other authors declare no competing financial interests.
Figures





References
-
- Beemsterboer SN, Homburg R, Gorter NA, Schats R, Hompes PGA, Lambalk CB.. The paradox of declining fertility but increasing twinning rates with advancing maternal age. Hum Reprod 2006;21:1531–1532. - PubMed
-
- Boomsma DI, Willemsen G, Sullivan PF, Heutink P, Meijer P, Sondervan D, Kluft C, Smit G, Nolen W a, Zitman FG. et al. Genome-wide association of major depression: description of samples for the GAIN Major Depressive Disorder Study: NTR and NESDA biobank projects. Eur J Hum Genet 2008;16:335–342. - PubMed
-
- Bulik-Sullivan BK, Loh P-R, Finucane HK, Ripke S, Yang J, Patterson N, Daly MJ, Price AL, Neale BM; Schizophrenia Working Group of the Psychiatric Genomics Consortium. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 2015;47:291–295. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R37 AA009367/AA/NIAAA NIH HHS/United States
- R01 AA009367/AA/NIAAA NIH HHS/United States
- RC2 MH089995/MH/NIMH NIH HHS/United States
- R01 HL043851/HL/NHLBI NIH HHS/United States
- AA09367/AA/NIAAA NIH HHS/United States
- NIH R01 HD042157-01A1/NH/NIH HHS/United States
- 230374/ERC_/European Research Council/International
- MRC_/Medical Research Council/United Kingdom
- K05 AA000145/AA/NIAAA NIH HHS/United States
- R01 CA047988/CA/NCI NIH HHS/United States
- RC2 MH089951/MH/NIMH NIH HHS/United States
- R01 DA005147/DA/NIDA NIH HHS/United States
- ERC-230374/ERC_/European Research Council/International
- R01 MH081802/MH/NIMH NIH HHS/United States
- R01 HD042157/HD/NICHD NIH HHS/United States
- R01 AA011886/AA/NIAAA NIH HHS/United States
- DA05147/DA/NIDA NIH HHS/United States
- R01 AG046938/AG/NIA NIH HHS/United States
- R01 AA009203/AA/NIAAA NIH HHS/United States
- U01 DA024417/DA/NIDA NIH HHS/United States
- U24 MH068457-06/MH/NIMH NIH HHS/United States
- R01 DA024417/DA/NIDA NIH HHS/United States
- ROI D0042157-01A/NH/NIH HHS/United States
- K02 AA018755/AA/NIAAA NIH HHS/United States
- U24 MH068457/MH/NIMH NIH HHS/United States
- UM1 CA182913/CA/NCI NIH HHS/United States
- R01 AA012502/AA/NIAAA NIH HHS/United States
- R01 HL080467/HL/NHLBI NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- HL043851/HL/NHLBI NIH HHS/United States
- R01 AA015416/AA/NIAAA NIH HHS/United States
- R37 DA005147/DA/NIDA NIH HHS/United States
- CA047988/CA/NCI NIH HHS/United States
- R01 DA013240/DA/NIDA NIH HHS/United States
- R37 AA012502/AA/NIAAA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

