Reverse metabolomics for the discovery of chemical structures from humans
- PMID: 38052229
- PMCID: PMC10849969
- DOI: 10.1038/s41586-023-06906-8
Reverse metabolomics for the discovery of chemical structures from humans
Abstract
Determining the structure and phenotypic context of molecules detected in untargeted metabolomics experiments remains challenging. Here we present reverse metabolomics as a discovery strategy, whereby tandem mass spectrometry spectra acquired from newly synthesized compounds are searched for in public metabolomics datasets to uncover phenotypic associations. To demonstrate the concept, we broadly synthesized and explored multiple classes of metabolites in humans, including N-acyl amides, fatty acid esters of hydroxy fatty acids, bile acid esters and conjugated bile acids. Using repository-scale analysis1,2, we discovered that some conjugated bile acids are associated with inflammatory bowel disease (IBD). Validation using four distinct human IBD cohorts showed that cholic acids conjugated to Glu, Ile/Leu, Phe, Thr, Trp or Tyr are increased in Crohn's disease. Several of these compounds and related structures affected pathways associated with IBD, such as interferon-γ production in CD4+ T cells3 and agonism of the pregnane X receptor4. Culture of bacteria belonging to the Bifidobacterium, Clostridium and Enterococcus genera produced these bile amidates. Because searching repositories with tandem mass spectrometry spectra has only recently become possible, this reverse metabolomics approach can now be used as a general strategy to discover other molecules from human and animal ecosystems.
© 2023. The Author(s).
Conflict of interest statement
P.C.D. is on the scientific advisory board of Sirenas and Cybele Microbiome, and is founder and scientific advisor and has equity in Ometa Labs, Arome and Enveda (with approval by UC San Diego). M.W. is a co-founder and had equity in Ometa Labs. R.K. Gencirq (stock and SAB member), DayTwo (consultant and SAB member), Cybele (stock and consultant), Biomesense (stock, consultant, SAB member), Micronoma (stock, SAB member, co-founder) and Biota (stock, co-founder). No competing interests exist for E.C.G., S.L.C., M.P., P.B.-F., A.K.S., M.C.T., H.L., S.Z., T.Y., J.A.-P., D.R.P., A.T.A., A.K.J., F.H., M.S.-N., H.V., A.N.A., B.B., A.H., N.V.C., F.J.G., C.B.C., R.J.X., H.C., E.S.B., A.D.P. and D.S.
Figures




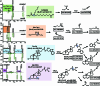
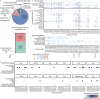

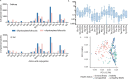
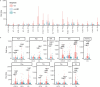


References
-
- Rovedatti, L. et al. Differential regulation of interleukin 17 and interferon γ production in inflammatory bowel disease. Gut58, 1629–1636 (2009). - PubMed
-
- Aksenov, A. A., da Silva, R., Knight, R., Lopes, N. P. & Dorrestein, P. C. Global chemical analysis of biology by mass spectrometry. Nat. Rev. Chem.1, 0054 (2017).
MeSH terms
Substances
Grants and funding
- RM1 GM145416/GM/NIGMS NIH HHS/United States
- P30 ES025128/ES/NIEHS NIH HHS/United States
- P30 DK040561/DK/NIDDK NIH HHS/United States
- R01 GM107550/GM/NIGMS NIH HHS/United States
- U01 DK119702/DK/NIDDK NIH HHS/United States
- P30 DK043351/DK/NIDDK NIH HHS/United States
- P41 GM103484/GM/NIGMS NIH HHS/United States
- P42 ES031009/ES/NIEHS NIH HHS/United States
- R01 DK136117/DK/NIDDK NIH HHS/United States
- R01 GM141277/GM/NIGMS NIH HHS/United States
- T32 DK007202/DK/NIDDK NIH HHS/United States
- P42 ES027704/ES/NIEHS NIH HHS/United States
- R00 DK110534/DK/NIDDK NIH HHS/United States
- R03 CA211211/CA/NCI NIH HHS/United States
- U24 DK133658/DK/NIDDK NIH HHS/United States
- K23 DK123406/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

