Nonsense-mediated mRNA decay, a simplified view of a complex mechanism
- PMID: 38052423
- PMCID: PMC10761751
- DOI: 10.5483/BMBRep.2023-0190
Nonsense-mediated mRNA decay, a simplified view of a complex mechanism
Abstract
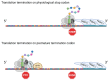
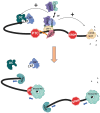
Nonsense-mediated mRNA decay (NMD) is both a quality control mechanism and a gene regulation pathway. It has been studied for more than 30 years, with an accumulation of many mechanistic details that have often led to debate and hence to different models of NMD activation, particularly in higher eukaryotes. Two models seem to be opposed, since the first requires intervention of the exon junction complex (EJC) to recruit NMD factors downstream of the premature termination codon (PTC), whereas the second involves an EJC-independent mechanism in which NMD factors concentrate in the 3'UTR to initiate NMD in the presence of a PTC. In this review we describe both models, giving recent molecular details and providing experimental arguments supporting one or the other model. In the end it is certainly possible to imagine that these two mechanisms co-exist, rather than viewing them as mutually exclusive. [BMB Reports 2023; 56(12): 625-632].
Conflict of interest statement
The authors have no conflicting interests.
Figures

Similar articles
-
Comparison of EJC-enhanced and EJC-independent NMD in human cells reveals two partially redundant degradation pathways.RNA. 2013 Oct;19(10):1432-48. doi: 10.1261/rna.038893.113. Epub 2013 Aug 20. RNA. 2013. PMID: 23962664 Free PMC article.
-
The role of nucleotide composition in premature termination codon recognition.BMC Bioinformatics. 2016 Dec 7;17(1):519. doi: 10.1186/s12859-016-1384-z. BMC Bioinformatics. 2016. PMID: 27927164 Free PMC article.
-
No-nonsense: insights into the functional interplay of nonsense-mediated mRNA decay factors.Biochem J. 2022 May 13;479(9):973-993. doi: 10.1042/BCJ20210556. Biochem J. 2022. PMID: 35551602 Free PMC article. Review.
-
Mechanism of Nonsense-Mediated mRNA Decay Stimulation by Splicing Factor SRSF1.Cell Rep. 2018 May 15;23(7):2186-2198. doi: 10.1016/j.celrep.2018.04.039. Cell Rep. 2018. PMID: 29768215 Free PMC article.
-
The Branched Nature of the Nonsense-Mediated mRNA Decay Pathway.Trends Genet. 2021 Feb;37(2):143-159. doi: 10.1016/j.tig.2020.08.010. Epub 2020 Sep 29. Trends Genet. 2021. PMID: 33008628 Free PMC article. Review.
Cited by
-
The NEDD4-binding protein N4BP1 degrades mRNA substrates through the coding sequence independent of nonsense-mediated decay.J Biol Chem. 2024 Dec;300(12):107954. doi: 10.1016/j.jbc.2024.107954. Epub 2024 Nov 2. J Biol Chem. 2024. PMID: 39491646 Free PMC article.
-
Trypanosomes lack a canonical EJC but possess an UPF1 dependent NMD-like pathway.PLoS One. 2025 Mar 7;20(3):e0315659. doi: 10.1371/journal.pone.0315659. eCollection 2025. PLoS One. 2025. PMID: 40053537 Free PMC article.
-
Knockdown of Gene Expression Following Activation of the NMD Pathway Using Splice-Switching Oligonucleotides.Methods Mol Biol. 2025;2962:237-253. doi: 10.1007/978-1-0716-4726-4_16. Methods Mol Biol. 2025. PMID: 40699433
-
Human FCHO1 deficiency: review of the literature and additional two cases.Clin Exp Immunol. 2025 Jan 21;219(1):uxae097. doi: 10.1093/cei/uxae097. Clin Exp Immunol. 2025. PMID: 39498505 Review.
-
PYM1 limits non-canonical Exon Junction Complex occupancy in a gene architecture dependent manner to tune mRNA expression.bioRxiv [Preprint]. 2025 Mar 15:2025.03.13.643037. doi: 10.1101/2025.03.13.643037. bioRxiv. 2025. PMID: 40161626 Free PMC article. Preprint.

