Enhanced production of mesencephalic dopaminergic neurons from lineage-restricted human undifferentiated stem cells
- PMID: 38052784
- PMCID: PMC10698156
- DOI: 10.1038/s41467-023-43471-0
Enhanced production of mesencephalic dopaminergic neurons from lineage-restricted human undifferentiated stem cells
Abstract
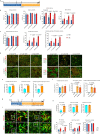
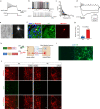
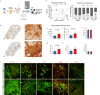
Current differentiation protocols for generating mesencephalic dopaminergic (mesDA) neurons from human pluripotent stem cells result in grafts containing only a small proportion of mesDA neurons when transplanted in vivo. In this study, we develop lineage-restricted undifferentiated stem cells (LR-USCs) from pluripotent stem cells, which enhances their potential for differentiating into caudal midbrain floor plate progenitors and mesDA neurons. Using a ventral midbrain protocol, 69% of LR-USCs become bona fide caudal midbrain floor plate progenitors, compared to only 25% of human embryonic stem cells (hESCs). Importantly, LR-USCs generate significantly more mesDA neurons under midbrain and hindbrain conditions in vitro and in vivo. We demonstrate that midbrain-patterned LR-USC progenitors transplanted into 6-hydroxydopamine-lesioned rats restore function in a clinically relevant non-pharmacological behavioral test, whereas midbrain-patterned hESC-derived progenitors do not. This strategy demonstrates how lineage restriction can prevent the development of undesirable lineages and enhance the conditions necessary for mesDA neuron generation.
© 2023. The Author(s).
Conflict of interest statement
M.M. and M.D. are inventors on a patent application filed by Aarhus University, WO2022136306A1 (National phase) that is related to the generation of lineage restricted cells. J.C.N. and J.R.C. are employed by Novo Nordisk. The remaining authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

