Functional divergence of a bacterial enzyme promotes healthy or acneic skin
- PMID: 38052825
- PMCID: PMC10697930
- DOI: 10.1038/s41467-023-43833-8
Functional divergence of a bacterial enzyme promotes healthy or acneic skin
Abstract
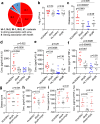
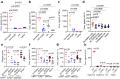
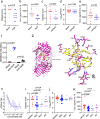
Acne is a dermatologic disease with a strong pathologic association with human commensal Cutibacterium acnes. Conspicuously, certain C. acnes phylotypes are associated with acne, whereas others are associated with healthy skin. Here we investigate if the evolution of a C. acnes enzyme contributes to health or acne. Two hyaluronidase variants exclusively expressed by C. acnes strains, HylA and HylB, demonstrate remarkable clinical correlation with acne or health. We show that HylA is strongly pro-inflammatory, and HylB is modestly anti-inflammatory in a murine (female) acne model. Structural and phylogenic studies suggest that the enzymes evolved from a common hyaluronidase that acquired distinct enzymatic activity. Health-associated HylB degrades hyaluronic acid (HA) exclusively to HA disaccharides leading to reduced inflammation, whereas HylA generates large-sized HA fragments that drive robust TLR2-dependent pathology. Replacing an amino acid, Serine to Glycine near the HylA catalytic site enhances the enzymatic activity of HylA and produces an HA degradation pattern intermediate to HylA and HylB. Selective targeting of HylA using peptide vaccine or inhibitors alleviates acne pathology. We suggest that the functional divergence of HylA and HylB is a major driving force behind C. acnes health- and acne- phenotype and propose targeting of HylA as an approach for acne therapy.
© 2023. The Author(s).
Conflict of interest statement
R.M., G.Y.L., I.A.H. and S.K. have filed for a patent application for the use of selective HylA inhibitors and vaccines as therapeutics. The remaining authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

