Shuttle peptide delivers base editor RNPs to rhesus monkey airway epithelial cells in vivo
- PMID: 38052872
- PMCID: PMC10698009
- DOI: 10.1038/s41467-023-43904-w
Shuttle peptide delivers base editor RNPs to rhesus monkey airway epithelial cells in vivo
Abstract
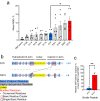
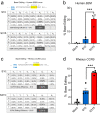
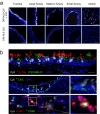
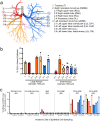
Gene editing strategies for cystic fibrosis are challenged by the complex barrier properties of airway epithelia. We previously reported that the amphiphilic S10 shuttle peptide non-covalently combined with CRISPR-associated (Cas) ribonucleoprotein (RNP) enabled editing of human and mouse airway epithelial cells. Here, we derive the S315 peptide as an improvement over S10 in delivering base editor RNP. Following intratracheal aerosol delivery of Cy5-labeled peptide in rhesus macaques, we confirm delivery throughout the respiratory tract. Subsequently, we target CCR5 with co-administration of ABE8e-Cas9 RNP and S315. We achieve editing efficiencies of up-to 5.3% in rhesus airway epithelia. Moreover, we document persistence of edited epithelia for up to 12 months in mice. Finally, delivery of ABE8e-Cas9 targeting the CFTR R553X mutation restores anion channel function in cultured human airway epithelia. These results demonstrate the therapeutic potential of base editor delivery with S315 to functionally correct the CFTR R553X mutation in respiratory epithelia.
© 2023. The Author(s).
Conflict of interest statement
P.B.M. is on the SAB and performs sponsored research for Spirovant Sciences, Inc. D.G. holds equity in Feldan Therapeutics. X.C., M.H. and D.G. are employees of Feldan Therapeutics. S.H., H.B.F. and J.R. were employees of Feldan Therapeutics. D.G. is co-inventor on patents and patent applications filed by Feldan Bio Inc. on the Shuttle peptide technology. D.R.L. is a consultant and equity holder of Beam Therapeutics, Prime Medicine, Pairwise Plants, Chroma Medicine, and Nvelop Therapeutics, companies that use or deliver gene editing or epigenome modulating agents. D.R.L. and G.A.N. are co-inventors on patent applications filed by the Broad Institute on base editing and its applications. The remaining authors declare no competing interests.
Figures


Update of
-
Shuttle Peptide Delivers Base Editor RNPs to Rhesus Monkey Airway Epithelial Cells In Vivo.Res Sq [Preprint]. 2023 Feb 17:rs.3.rs-2540755. doi: 10.21203/rs.3.rs-2540755/v1. Res Sq. 2023. Update in: Nat Commun. 2023 Dec 5;14(1):8051. doi: 10.1038/s41467-023-43904-w. PMID: 36824928 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
- UG3 AI150551/AI/NIAID NIH HHS/United States
- K99 HL163805/HL/NHLBI NIH HHS/United States
- P01 HL152960/HL/NHLBI NIH HHS/United States
- R00 HL163805/HL/NHLBI NIH HHS/United States
- UG3 HL147366/HL/NHLBI NIH HHS/United States
- R35 GM118062/GM/NIGMS NIH HHS/United States
- S10 RR025063/RR/NCRR NIH HHS/United States
- U01 AI142756/AI/NIAID NIH HHS/United States
- P30 DK054759/DK/NIDDK NIH HHS/United States
- S10 OD028713/OD/NIH HHS/United States
- S10 OD016261/OD/NIH HHS/United States
- P30 ES005605/ES/NIEHS NIH HHS/United States
- RM1 HG009490/HG/NHGRI NIH HHS/United States
- P51 OD011107/OD/NIH HHS/United States
- U24 HG010423/HG/NHGRI NIH HHS/United States
- U42 OD027094/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

