Pseudorabies virus upregulates low-density lipoprotein receptors to facilitate viral entry
- PMID: 38054618
- PMCID: PMC10804996
- DOI: 10.1128/jvi.01664-23
Pseudorabies virus upregulates low-density lipoprotein receptors to facilitate viral entry
Abstract
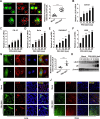
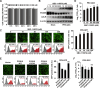
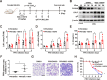
Pseudorabies virus (PRV) is the causative agent of Aujeszky's disease in pigs. The low-density lipoprotein receptor (LDLR) is a transcriptional target of the sterol-regulatory element-binding proteins (SREBPs) and participates in the uptake of LDL-derived cholesterol. However, the involvement of LDLR in PRV infection has not been well characterized. We observed an increased expression level of LDLR mRNA in PRV-infected 3D4/21, PK-15, HeLa, RAW264.7, and L929 cells. The LDLR protein level was also upregulated by PRV infection in PK-15 cells and in murine lung and brain. The treatment of cells with the SREBP inhibitor, fatostatin, or with SREBP2-specific small interfering RNA prevented the PRV-induced upregulation of LDLR expression as well as viral protein expression and progeny virus production. This suggested that PRV activated SREBPs to induce LDLR expression. Furthermore, interference in LDLR expression affected PRV proliferation, while LDLR overexpression promoted it. This indicated that LDLR was involved in PRV infection. The study also demonstrated that LDLR participated in PRV invasions. The overexpression of LDLR or inhibition of proprotein convertase subtilisin/kexin type 9 (PCSK9), which binds to LDLR and targets it for lysosomal degradation, significantly enhanced PRV attachment and entry. Mechanistically, LDLR interacted with PRV on the plasma membrane, and pretreatment of cells with LDLR antibodies was able to neutralize viral entry. An in vivo study indicated that the treatment of mice with the PCSK9 inhibitor SBC-115076 promoted PRV proliferation. The data from the study indicate that PRV hijacks LDLR for viral entry through the activation of SREBPs.IMPORTANCEPseudorabies virus (PRV) is a herpesvirus that primarily manifests as fever, pruritus, and encephalomyelitis in various domestic and wild animals. Owing to its lifelong latent infection characteristics, PRV outbreaks have led to significant financial setbacks in the global pig industry. There is evidence that PRV variant strains can infect humans, thereby crossing the species barrier. Therefore, gaining deeper insights into PRV pathogenesis and developing updated strategies to contain its spread are critical. This study posits that the low-density lipoprotein receptor (LDLR) could be a co-receptor for PRV infection. Hence, strategies targeting LDLR may provide a promising avenue for the development of effective PRV vaccines and therapeutic interventions.
Keywords: LDLR; PCSK9; SREBPs; pseudorabies virus; viral entry.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

