Citrullination profile analysis reveals peptidylarginine deaminase 3 as an HSV-1 target to dampen the activity of candidate antiviral restriction factors
- PMID: 38055760
- PMCID: PMC10727434
- DOI: 10.1371/journal.ppat.1011849
Citrullination profile analysis reveals peptidylarginine deaminase 3 as an HSV-1 target to dampen the activity of candidate antiviral restriction factors
Abstract
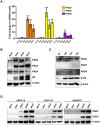
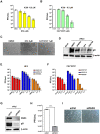
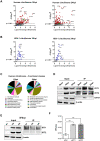
Herpes simplex virus 1 (HSV-1) is a neurotropic virus that remains latent in neuronal cell bodies but reactivates throughout an individual's life, causing severe adverse reactions, such as herpes simplex encephalitis (HSE). Recently, it has also been implicated in the etiology of Alzheimer's disease (AD). The absence of an effective vaccine and the emergence of numerous drug-resistant variants have called for the development of new antiviral agents that can tackle HSV-1 infection. Host-targeting antivirals (HTAs) have recently emerged as promising antiviral compounds that act on host-cell factors essential for viral replication. Here we show that a new class of HTAs targeting peptidylarginine deiminases (PADs), a family of calcium-dependent enzymes catalyzing protein citrullination, exhibits a marked inhibitory activity against HSV-1. Furthermore, we show that HSV-1 infection leads to enhanced protein citrullination through transcriptional activation of three PAD isoforms: PAD2, PAD3, and PAD4. Interestingly, PAD3-depletion by specific drugs or siRNAs dramatically inhibits HSV-1 replication. Finally, an analysis of the citrullinome reveals significant changes in the deimination levels of both cellular and viral proteins, with the interferon (IFN)-inducible proteins IFIT1 and IFIT2 being among the most heavily deiminated ones. As genetic depletion of IFIT1 and IFIT2 strongly enhances HSV-1 growth, we propose that viral-induced citrullination of IFIT1 and 2 is a highly efficient HSV-1 evasion mechanism from host antiviral resistance. Overall, our findings point to a crucial role of citrullination in subverting cellular responses to viral infection and demonstrate that PAD inhibitors efficiently suppress HSV-1 infection in vitro, which may provide the rationale for their repurposing as HSV-1 antiviral drugs.
Copyright: © 2023 Pasquero et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

