RAD54L2-mediated DNA damage avoidance pathway specifically preserves genome integrity in response to topoisomerase 2 poisons
- PMID: 38055811
- PMCID: PMC10699775
- DOI: 10.1126/sciadv.adi6681
RAD54L2-mediated DNA damage avoidance pathway specifically preserves genome integrity in response to topoisomerase 2 poisons
Abstract
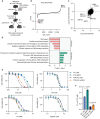
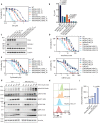
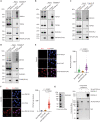
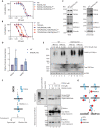
Type II topoisomerases (TOP2) form transient TOP2 cleavage complexes (TOP2ccs) during their catalytic cycle to relieve topological stress. TOP2ccs are covalently linked TOP2-DNA intermediates that are reversible but can be trapped by TOP2 poisons. Trapped TOP2ccs block transactions on DNA and generate genotoxic stress, which are the mechanisms of action of TOP2 poisons. How cells avoid TOP2cc accumulation remains largely unknown. In this study, we uncovered RAD54 like 2 (RAD54L2) as a key factor that mediates a TOP2-specific DNA damage avoidance pathway. RAD54L2 deficiency conferred unique sensitivity to treatment with TOP2 poisons. RAD54L2 interacted with TOP2A/TOP2B and ZATT/ZNF451 and promoted the turnover of TOP2 from DNA with or without TOP2 poisons. Additionally, inhibition of proteasome activity enhanced the chromatin binding of RAD54L2, which in turn led to the removal of TOP2 from chromatin. In conclusion, we propose that RAD54L2-mediated TOP2 turnover is critically important for the avoidance of potential TOP2-linked DNA damage under physiological conditions and in response to TOP2 poisons.
Figures



References
-
- J. J. Champoux, DNA topoisomerases: Structure, function, and mechanism. Annu. Rev. Biochem. 70, 369–413 (2001). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

