Innate TCRβ-chain engagement drives human T cells toward distinct memory-like effector phenotypes with immunotherapeutic potentials
- PMID: 38055824
- PMCID: PMC10699787
- DOI: 10.1126/sciadv.adj6174
Innate TCRβ-chain engagement drives human T cells toward distinct memory-like effector phenotypes with immunotherapeutic potentials
Abstract
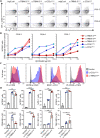
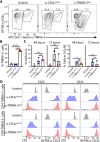
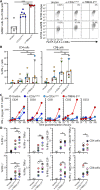
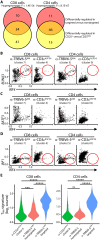
Clonotypic αβ T cell responses to cargoes presented by major histocompatibility complex (MHC), MR1, or CD1 proteins underpin adaptive immunity. Those responses are mostly mediated by complementarity-determining region 3 motifs created by quasi-random T cell receptor (TCR) gene rearrangements, with diversity being highest for TCRγδ. Nonetheless, TCRγδ also displays nonclonotypic innate responsiveness following engagement of germline-encoded Vγ-specific residues by butyrophilin (BTN) or BTN-like (BTNL) proteins that uniquely mediate γδ T cell subset selection. We now report that nonclonotypic TCR engagement likewise induces distinct phenotypes in TCRαβ+ cells. Specifically, antibodies to germline-encoded human TCRVβ motifs consistently activated naïve or memory T cells toward core states distinct from those induced by anti-CD3 or superantigens and from others commonly reported. Those states combined selective proliferation and effector function with activation-induced inhibitory receptors and memory differentiation. Thus, nonclonotypic TCRVβ targeting broadens our perspectives on human T cell response modes and might offer ways to induce clinically beneficial phenotypes in defined T cell subsets.
Figures



References
-
- Sette A., Sidney J., Crotty S., T Cell Responses to SARS-CoV-2. Annu. Rev. Immunol. 41, 343–373 (2023). - PubMed
-
- Mahnke Y. D., Brodie T. M., Sallusto F., Roederer M., Lugli E., The who's who of T-cell differentiation: Human memory T-cell subsets. Eur. J. Immunol. 43, 2797–2809 (2013). - PubMed
-
- Osman M., Park S. L., Mackay L. K., Tissue-resident memory T (TRM) cells: Front-line workers of the immune system. Eur. J. Immunol. 53, e2250060 (2023). - PubMed
-
- Alfei F., Kanev K., Hofmann M., Wu M., Ghoneim H. E., Roelli P., Utzschneider D. T., von Hoesslin M., Cullen J. G., Fan Y., Eisenberg V., Wohlleber D., Steiger K., Merkler D., Delorenzi M., Knolle P. A., Cohen C. J., Thimme R., Youngblood B., Zehn D., TOX reinforces the phenotype and longevity of exhausted T cells in chronic viral infection. Nature 571, 265–269 (2019). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

