Mitochondrial metabolic flexibility is critical for CD8+ T cell antitumor immunity
- PMID: 38055827
- PMCID: PMC10699783
- DOI: 10.1126/sciadv.adf9522
Mitochondrial metabolic flexibility is critical for CD8+ T cell antitumor immunity
Abstract
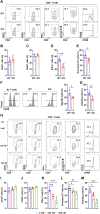
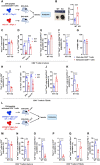
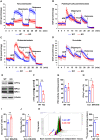
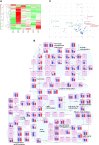
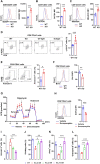
Mitochondria use different substrates for energy production and intermediatory metabolism according to the availability of nutrients and oxygen levels. The role of mitochondrial metabolic flexibility for CD8+ T cell immune response is poorly understood. Here, we report that the deletion or pharmacological inhibition of protein tyrosine phosphatase, mitochondrial 1 (PTPMT1) significantly decreased CD8+ effector T cell development and clonal expansion. In addition, PTPMT1 deletion impaired stem-like CD8+ T cell maintenance and accelerated CD8+ T cell exhaustion/dysfunction, leading to aggravated tumor growth. Mechanistically, the loss of PTPMT1 critically altered mitochondrial fuel selection-the utilization of pyruvate, a major mitochondrial substrate derived from glucose-was inhibited, whereas fatty acid utilization was enhanced. Persistent mitochondrial substrate shift and metabolic inflexibility induced oxidative stress, DNA damage, and apoptosis in PTPMT1 knockout cells. Collectively, this study reveals an important role of PTPMT1 in facilitating mitochondrial utilization of carbohydrates and that mitochondrial flexibility in energy source selection is critical for CD8+ T cell antitumor immunity.
Figures




References
-
- M. Hashimoto, A. O. Kamphorst, S. J. Im, H. T. Kissick, R. N. Pillai, S. S. Ramalingam, K. Araki, R. Ahmed, CD8 T cell exhaustion in chronic infection and cancer: Opportunities for interventions. Annu. Rev. Med. 69, 301–318 (2018). - PubMed
-
- S. J. Im, M. Hashimoto, M. Y. Gerner, J. Lee, H. T. Kissick, M. C. Burger, Q. Shan, J. S. Hale, J. Lee, T. H. Nasti, A. H. Sharpe, G. J. Freeman, R. N. Germain, H. I. Nakaya, H. H. Xue, R. Ahmed, Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 537, 417–421 (2016). - PMC - PubMed
-
- C. S. Eberhardt, H. T. Kissick, M. R. Patel, M. A. Cardenas, N. Prokhnevska, R. C. Obeng, T. H. Nasti, C. C. Griffith, S. J. Im, X. Wang, D. M. Shin, M. Carrington, Z. G. Chen, J. Sidney, A. Sette, N. F. Saba, A. Wieland, R. Ahmed, Functional HPV-specific PD-1+ stem-like CD8 T cells in head and neck cancer. Nature 597, 279–284 (2021). - PMC - PubMed
-
- D. T. Utzschneider, M. Charmoy, V. Chennupati, L. Pousse, D. P. Ferreira, S. Calderon-Copete, M. Danilo, F. Alfei, M. Hofmann, D. Wieland, S. Pradervand, R. Thimme, D. Zehn, W. Held, T cell factor 1-expressing memory-like CD8+ T cells sustain the immune response to chronic viral infections. Immunity 45, 415–427 (2016). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

