Duffy antigen is expressed during erythropoiesis in Duffy-negative individuals
- PMID: 38056457
- PMCID: PMC10843566
- DOI: 10.1016/j.chom.2023.10.019
Duffy antigen is expressed during erythropoiesis in Duffy-negative individuals
Abstract
The erythrocyte silent Duffy blood group phenotype in Africans is thought to confer resistance to Plasmodium vivax blood-stage infection. However, recent studies report P. vivax infections across Africa in Fy-negative individuals. This suggests that the globin transcription factor 1 (GATA-1) SNP underlying Fy negativity does not entirely abolish Fy expression or that P. vivax has developed a Fy-independent red blood cell (RBC) invasion pathway. We show that RBCs and erythroid progenitors from in vitro differentiated CD34 cells and from bone marrow aspirates from Fy-negative samples express a functional Fy on their surface. This suggests that the GATA-1 SNP does not entirely abolish Fy expression. Given these results, we developed an in vitro culture system for P. vivax and show P. vivax can invade erythrocytes from Duffy-negative individuals. This study provides evidence that Fy is expressed in Fy-negative individuals and explains their susceptibility to P. vivax with major implications and challenges for P. vivax malaria eradication.
Keywords: Duffy protein; Duffy-negative individuals; Plasmodium vivax invasion; bone marrow; erythroid progenitors.
Copyright © 2023. Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



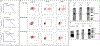

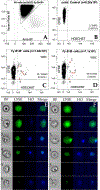
Comment in
-
Vivax malaria and Duffy antigen: Stop being so negative.Cell Host Microbe. 2023 Dec 13;31(12):1959-1960. doi: 10.1016/j.chom.2023.11.017. Cell Host Microbe. 2023. PMID: 38096790
References
-
- Bachelerie F, Ben-Baruch A, Burkhardt AM, Combadiere C, Farber JM, Graham GJ, Horuk R, Sparre-Ulrich AH, Locati M, Luster AD, et al. (2014). International Union of Pharmacology. LXXXIX. Update on the Extended Family of Chemokine Receptors and Introducing a New Nomenclature for Atypical Chemokine Receptors. Pharmacol Rev 66, 1–79. 10.1124/pr.113.007724. - DOI - PMC - PubMed
-
- Dawson TC, Lentsch AB, Wang Z, Cowhig JE, Rot A, Maeda N, and Peiper SC (2000). Exaggerated response to endotoxin in mice lacking the Duffy antigen/receptor for chemokines (DARC). Blood 96, 1681–1684. - PubMed
-
- Neote K, Darbonne W, Ogez J, Horuk R, and Schall TJ (1993). Identification of a promiscuous inflammatory peptide receptor on the surface of red blood cells. J Biol Chem 268, 12247–12249. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

