Systemically administered wound-homing peptide accelerates wound healing by modulating syndecan-4 function
- PMID: 38057316
- PMCID: PMC10700342
- DOI: 10.1038/s41467-023-43848-1
Systemically administered wound-homing peptide accelerates wound healing by modulating syndecan-4 function
Erratum in
-
Author Correction: Systemically administered wound-homing peptide accelerates wound healing by modulating syndecan-4 function.Nat Commun. 2024 Jan 3;15(1):234. doi: 10.1038/s41467-023-44574-4. Nat Commun. 2024. PMID: 38172142 Free PMC article. No abstract available.
Abstract
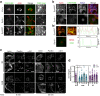
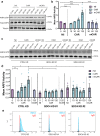
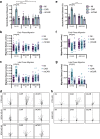
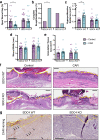
CAR (CARSKNKDC) is a wound-homing peptide that recognises angiogenic neovessels. Here we discover that systemically administered CAR peptide has inherent ability to promote wound healing: wounds close and re-epithelialise faster in CAR-treated male mice. CAR promotes keratinocyte migration in vitro. The heparan sulfate proteoglycan syndecan-4 regulates cell migration and is crucial for wound healing. We report that syndecan-4 expression is restricted to epidermis and blood vessels in mice skin wounds. Syndecan-4 regulates binding and internalisation of CAR peptide and CAR-mediated cytoskeletal remodelling. CAR induces syndecan-4-dependent activation of the small GTPase ARF6, via the guanine nucleotide exchange factor cytohesin-2, and promotes syndecan-4-, ARF6- and Cytohesin-2-mediated keratinocyte migration. Finally, we show that genetic ablation of syndecan-4 in male mice eliminates CAR-induced wound re-epithelialisation following systemic administration. We propose that CAR peptide activates syndecan-4 functions to selectively promote re-epithelialisation. Thus, CAR peptide provides a therapeutic approach to enhance wound healing in mice; systemic, yet target organ- and cell-specific.
© 2023. The Author(s).
Conflict of interest statement
E.R. has ownership interest (including patents) in Vascular Biosciences Inc., biotech company developing the CAR peptide for clinical applications. No conflicts of interest were disclosed by the other authors.
Figures






References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

