Comparative and phylogenetic analysis of Chiloschista (Orchidaceae) species and DNA barcoding investigation based on plastid genomes
- PMID: 38057701
- PMCID: PMC10702055
- DOI: 10.1186/s12864-023-09847-8
Comparative and phylogenetic analysis of Chiloschista (Orchidaceae) species and DNA barcoding investigation based on plastid genomes
Abstract
Background: Chiloschista (Orchidaceae, Aeridinae) is an epiphytic leafless orchid that is mainly distributed in tropical or subtropical forest canopies. This rare and threatened orchid lacks molecular resources for phylogenetic and barcoding analysis. Therefore, we sequenced and assembled seven complete plastomes of Chiloschista to analyse the plastome characteristics and phylogenetic relationships and conduct a barcoding investigation.
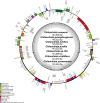
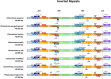
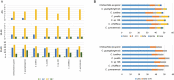
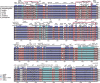
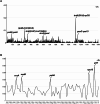
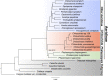
Results: We are the first to publish seven Chiloschista plastomes, which possessed the typical quadripartite structure and ranged from 143,233 bp to 145,463 bp in size. The plastomes all contained 120 genes, consisting of 74 protein-coding genes, 38 tRNA genes and eight rRNA genes. The ndh genes were pseudogenes or lost in the genus, and the genes petG and psbF were under positive selection. The seven Chiloschista plastomes displayed stable plastome structures with no large inversions or rearrangements. A total of 14 small inversions (SIs) were identified in the seven Chiloschista plastomes but were all similar within the genus. Six noncoding mutational hotspots (trnNGUU-rpl32 > rpoB-trnCGCA > psbK-psbI > psaC-rps15 > trnEUUC-trnTGGU > accD-psaI) and five coding sequences (ycf1 > rps15 > matK > psbK > ccsA) were selected as potential barcodes based on nucleotide diversity and species discrimination analysis, which suggested that the potential barcode ycf1 was most suitable for species discrimination. A total of 47-56 SSRs and 11-14 long repeats (> 20 bp) were identified in Chiloschista plastomes, and they were mostly located in the large single copy intergenic region. Phylogenetic analysis indicated that Chiloschista was monophyletic. It was clustered with Phalaenopsis and formed the basic clade of the subtribe Aeridinae with a moderate support value. The results also showed that seven Chiloschista species were divided into three major clades with full support.
Conclusion: This study was the first to analyse the plastome characteristics of the genus Chiloschista in Orchidaceae, and the results showed that Chiloschista plastomes have conserved plastome structures. Based on the plastome hotspots of nucleotide diversity, several genes and noncoding regions are suitable for phylogenetic and population studies. Chiloschista may provide an ideal system to investigate the dynamics of plastome evolution and DNA barcoding investigation for orchid studies.
Keywords: Aeridinae; Chloroplast genome; DNA barcoding; Phylogenetic analysis; Small inversions.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Zhang L, Huang Y, Huang J, Ya J, Zhe M, Zeng C, et al. DNA barcoding of Cymbidium by genome skimming: call for next-generation nuclear barcodes. Mol Ecol Resour. 2022;23:1–16. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

