Immune checkpoints are predominantly co-expressed by clonally expanded CD4+FoxP3+ intratumoral T-cells in primary human cancers
- PMID: 38057799
- PMCID: PMC10699039
- DOI: 10.1186/s13046-023-02897-6
Immune checkpoints are predominantly co-expressed by clonally expanded CD4+FoxP3+ intratumoral T-cells in primary human cancers
Abstract
Background: In addition to anti-PD(L)1, anti-CTLA-4 and anti-LAG-3, novel immune checkpoint proteins (ICP)-targeted antibodies have recently failed to demonstrate significant efficacy in clinical trials. In these trials, patients were enrolled without screening for drug target expression. Although these novel ICP-targeted antibodies were expected to stimulate anti-tumor CD8 + T-cells, the rationale for their target expression in human tumors relied on pre-clinical IHC stainings and transcriptomic data, which are poorly sensitive and specific techniques for assessing membrane protein expression on immune cell subsets. Our aim was to describe ICP expression on intratumoral T-cells from primary solid tumors to better design upcoming neoadjuvant cancer immunotherapy trials.
Methods: We prospectively performed multiparameter flow cytometry and single-cell RNA sequencing (scRNA-Seq) paired with TCR sequencing on freshly resected human primary tumors of various histological types to precisely determine ICP expression levels within T-cell subsets.
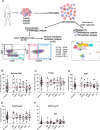
Results: Within a given tumor type, we found high inter-individual variability for tumor infiltrating CD45 + cells and for T-cells subsets. The proportions of CD8+ T-cells (~ 40%), CD4+ FoxP3- T-cells (~ 40%) and CD4+ FoxP3+ T-cells (~ 10%) were consistent across patients and indications. Intriguingly, both stimulatory (CD25, CD28, 4-1BB, ICOS, OX40) and inhibitory (PD-1, CTLA-4, PD-L1, CD39 and TIGIT) checkpoint proteins were predominantly co-expressed by intratumoral CD4+FoxP3+ T-cells. ScRNA-Seq paired with TCR sequencing revealed that T-cells with high clonality and high ICP expressions comprised over 80% of FoxP3+ cells among CD4+ T-cells. Unsupervised clustering of flow cytometry and scRNAseq data identified subsets of CD8+ T-cells and of CD4+ FoxP3- T-cells expressing certain checkpoints, though these expressions were generally lower than in CD4+ FoxP3+ T-cell subsets, both in terms of proportions among total T-cells and ICP expression levels.
Conclusions: Tumor histology alone does not reveal the complete picture of the tumor immune contexture. In clinical trials, assumptions regarding target expression should rely on more sensitive and specific techniques than conventional IHC or transcriptomics. Flow cytometry and scRNAseq accurately characterize ICP expression within immune cell subsets. Much like in hematology, flow cytometry can better describe the immune contexture of solid tumors, offering the opportunity to guide patient treatment according to drug target expression rather than tumor histological type.
Keywords: Cancer; Flow cytometry; Immune checkpoints; Immunology; Immunotherapy; Single-cell RNA-Seq; T-cells; TCR repertoire.
© 2023. The Author(s).
Conflict of interest statement
This project was funded by the Fondation MSD Avenir (
Figures




References
-
- Bendell JC, Bedard P, Bang YJ, LoRusso P, Hodi S, Gordon M, et al. Abstract CT302: Phase Ia/Ib dose-escalation study of the anti-TIGIT antibody tiragolumab as a single agent and in combination with atezolizumab in patients with advanced solid tumors. Cancer Res. 2020;80(16_Supplement):CT302. doi: 10.1158/1538-7445.AM2020-CT302. - DOI
-
- Niu J, Maurice-Dror C, Lee DH, Kim DW, Nagrial A, Voskoboynik M, et al. First-in-human phase 1 study of the anti-TIGIT antibody vibostolimab as monotherapy or with pembrolizumab for advanced solid tumors, including non-small-cell lung cancer☆. Ann Oncol Off J Eur Soc Med Oncol. 2022;33(2):169–80. doi: 10.1016/j.annonc.2021.11.002. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

