CircATP13A1 (hsa_circ_0000919) promotes cell proliferation and metastasis and inhibits cell apoptosis in pancreatic ductal adenocarcinoma via the miR-186/miR-326/HMGA2 axis: implications for novel therapeutic targets
- PMID: 38058810
- PMCID: PMC10695776
CircATP13A1 (hsa_circ_0000919) promotes cell proliferation and metastasis and inhibits cell apoptosis in pancreatic ductal adenocarcinoma via the miR-186/miR-326/HMGA2 axis: implications for novel therapeutic targets
Abstract
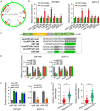
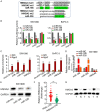
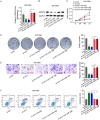
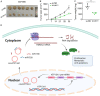
Pancreatic ductal adenocarcinoma (PDAC) is a notoriously aggressive malignancy with a survival rate of merely 9%. The prognosis in patients with PDAC is relatively poor, particularly in patients with advanced distant metastases. However, the mechanisms of PDAC progression remain elusive. Circular RNAs (circRNAs) have been implicated in the development of various malignancies, including PDAC. Therefore, this study aimed to investigate how a novel circRNA, circATP13A1, regulates PDAC progression. We used the GEO database to determine circATP13A1 expression levels in cancer and adjacent cells and employed the limma package of R software to identify differentially expressed circRNAs. We detected the expression of circATP13A1, miR-186, and miR-326 using qRT-PCR and investigated the effect of circATP13A1 on cell proliferation, migration, invasion, and apoptosis in vitro using the Cell Counting Kit-8 (CCK-8), the transwell migration assay, and the flow cytometry assay. We then performed RNA pull-down assay, RNA immunoprecipitation (RIP), and Western blot to verify the interaction between circATP13A1, miR-186, miR-326, and HMGA2. Moreover, we used a naked mice model to determine how circATP13A1 affects tumor growth and progression in vivo. Loss and gain of function analyses revealed that circATP13A1 upregulation promotes cell proliferation, migration, invasion and tumor growth both in vitro and in vivo, which results in PDAC progression and poor prognosis in patients. CircATP13A1 knockdown significantly impaired cell proliferation and migration of PDAC cell lines. Additionally, circATP13A1 knockdown significantly increased the expression of miR-186 and miR-326, while reducing the expression of HMGA2 (P < 0.05), indicating that miR-186 and miR-326 are downstream targets of circATP13A1. Rescue experiments support the interactions between circATP13A1, miR-186, miR-326, and HMGA2. In conclusion, we demonstrated that circATP13A1 sponges the miR-186/miR-326/HMGA2/axis, acting as an oncogene to promote PDAC development.
Keywords: CircATP13A1; HMGA2; PDAC; miR-186; miR-326.
AJCR Copyright © 2023.
Conflict of interest statement
None.
Figures


Similar articles
-
Circ_100565 promotes proliferation, migration and invasion in non-small cell lung cancer through upregulating HMGA2 via sponging miR-506-3p.Cancer Cell Int. 2020 May 12;20:160. doi: 10.1186/s12935-020-01241-8. eCollection 2020. Cancer Cell Int. 2020. PMID: 32425695 Free PMC article.
-
Novel circularRNA circ-0047078 promotes pancreatic ductal adenocarcinoma progression through mircoRNA miR-11181- Chemokine (C-X-C motif) Ligand 12/Melanoma Cell Adhesion Molecule/Regulator of G-protein signaling 16 pathway.Mol Biol Rep. 2022 Sep;49(9):8761-8775. doi: 10.1007/s11033-022-07723-4. Epub 2022 Jun 30. Mol Biol Rep. 2022. PMID: 35771357
-
Identifying a novel IRF3/circUHRF1/miR-1306-5p/ARL4C axis in pancreatic ductal adenocarcinoma progression.Cell Cycle. 2022 Feb;21(4):392-405. doi: 10.1080/15384101.2021.2020450. Epub 2022 Jan 5. Cell Cycle. 2022. PMID: 34983293 Free PMC article.
-
Exosome-transmitted hsa_circ_0012634 suppresses pancreatic ductal adenocarcinoma progression through regulating miR-147b/HIPK2 axis.Cancer Biol Ther. 2023 Dec 31;24(1):2218514. doi: 10.1080/15384047.2023.2218514. Cancer Biol Ther. 2023. PMID: 37326330 Free PMC article.
-
Upregulated circRNA hsa_circ_0071036 promotes tumourigenesis of pancreatic cancer by sponging miR-489 and predicts unfavorable characteristics and prognosis.Cell Cycle. 2021 Feb;20(4):369-382. doi: 10.1080/15384101.2021.1874684. Epub 2021 Jan 28. Cell Cycle. 2021. PMID: 33507122 Free PMC article.
Cited by
-
CircRNA hsa_circ_0004781 promoted cell proliferation by acting as a sponge for miR-9-5p and miR-338-3p and upregulating KLF5 and ADAM17 expression in pancreatic ductal adenocarcinoma.Cancer Cell Int. 2025 Feb 19;25(1):56. doi: 10.1186/s12935-025-03687-0. Cancer Cell Int. 2025. PMID: 39972489 Free PMC article.
-
CircMALAT1 promotes the proliferation and metastasis of intrahepatic cholangiocarcinoma via the miR-512-5p/VCAM1 axis.Acta Biochim Biophys Sin (Shanghai). 2024 Oct 23;57(2):223-236. doi: 10.3724/abbs.2024185. Acta Biochim Biophys Sin (Shanghai). 2024. PMID: 39463204 Free PMC article.
References
-
- Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. - PubMed
-
- Long J, Luo GP, Xiao ZW, Liu ZQ, Guo M, Liu L, Liu C, Xu J, Gao YT, Zheng Y, Wu C, Ni QX, Li M, Yu X. Cancer statistics: current diagnosis and treatment of pancreatic cancer in Shanghai, China. Cancer Lett. 2014;346:273–277. - PubMed
-
- Saif MW. Pancreatic cancer: highlights from the 42nd annual meeting of the American Society of Clinical Oncology, 2006. JOP. 2006;7:337–348. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
