IGF2BP3-mediated regulation of GLS and GLUD1 gene expression promotes treg-induced immune escape in human cervical cancer
- PMID: 38058838
- PMCID: PMC10695810
IGF2BP3-mediated regulation of GLS and GLUD1 gene expression promotes treg-induced immune escape in human cervical cancer
Abstract
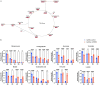
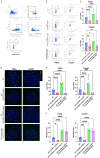
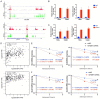
This study aimed to investigate the impact of IGF2BP3, a well-known m6A modification-related protein, on the metabolic and immune microenvironment of human cervical cancer. Bioinformatics analysis was performed to analyze the expression of IGF2BP3 in various databases, and its findings were validated using human cervical cancer tissue microarrays. We conducted a study to investigate the impact of IGF2BP3 on glutamine metabolism in cervical cancer cells through the application of metabolomics and metabolic flow analysis. Additionally, we explored how cervical cancer cells promote immune escape by secreting glutamine-derived lactate in a 3D culture setting. To identify the specific targets of IGF2BP3 that influence glutamine metabolism in cervical cancer, we employed RIP-seq analysis. IGF2BP3 exhibited high expression levels in multiple cervical cancer datasets, and its expression was significantly associated with the prognosis of cervical cancer patients. In mixed 3D cell cultures of cervical cancer and T cells, IGF2BP3 was found to enhance glutamate and glutamine metabolism in cervical cancer cells by up regulating the expression of GLS and GLUD1 genes. Moreover, it influenced the differentiation of Treg cells by promoting lactate production and secretion in cervical cancer, leading to immune escape. Mechanistic analysis revealed that IGF2BP3 stabilized the mRNA of GLS and GLUD1 genes through m6A modification, thereby facilitating glutamate and glutamine metabolism in cervical cancer cells and regulating lactate production. Additionally, we investigated the correlation between GLS, GLUD1 protein expression, and IGF2BP3 expression in human cervical cancer through multicolor immunofluorescence staining. The relevance of IGF2BP3 in the context of Treg cell-associated immune escape in cervical cancer was also confirmed. IGF2BP3 exhibits high expression in human cervical cancer and plays a crucial role in stabilizing the mRNA of GLS and GLUD1 genes, key metabolic enzymes in glutamate and glutamine metabolism, through m6A modification. This process leads to immune escape in cervical cancer by promoting lactate production and secretion.
Keywords: IGF2BP3; M6A modification; cervical cancer; glutamine metabolism.
AJCR Copyright © 2023.
Conflict of interest statement
None.
Figures




References
-
- Sendinc E, Shi Y. RNA m6A methylation across the transcriptome. Mol Cell. 2023;83:428–441. - PubMed
-
- Liu Y, Yang D, Liu T, Chen J, Yu J, Yi P. N6-methyladenosine-mediated gene regulation and therapeutic implications. Trends Mol Med. 2023;29:454–467. - PubMed
-
- Zhu X, Fu H, Sun J, Xu Q. Interaction between N6-methyladenosine (m6A) modification and environmental chemical-induced diseases in various organ systems. Chem Biol Interact. 2023;373:110376. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
