Additional PfCRT mutations driven by selective pressure for improved fitness can result in the loss of piperaquine resistance and altered Plasmodium falciparum physiology
- PMID: 38059639
- PMCID: PMC10790694
- DOI: 10.1128/mbio.01832-23
Additional PfCRT mutations driven by selective pressure for improved fitness can result in the loss of piperaquine resistance and altered Plasmodium falciparum physiology
Abstract
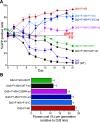
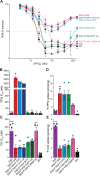
Our study leverages gene editing techniques in Plasmodium falciparum asexual blood stage parasites to profile novel mutations in mutant PfCRT, an important mediator of piperaquine resistance, which developed in Southeast Asian field isolates or in parasites cultured for long periods of time. We provide evidence that increased parasite fitness of these lines is the primary driver for the emergence of these PfCRT variants. These mutations differentially impact parasite susceptibility to piperaquine and chloroquine, highlighting the multifaceted effects of single point mutations in this transporter. Molecular features of drug resistance and parasite physiology were examined in depth using proteoliposome-based drug uptake studies and peptidomics, respectively. Energy minimization calculations, showing how these novel mutations might impact the PfCRT structure, suggested a small but significant effect on drug interactions. This study reveals the subtle interplay between antimalarial resistance, parasite fitness, PfCRT structure, and intracellular peptide availability in PfCRT-mediated parasite responses to changing drug selective pressures.
Keywords: PfCRT; Plasmodium falciparum; drug resistance evolution; fitness; malaria.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- World Health Organization . 2022. World malaria report. Available from: https://www.who.int/teams/global-malaria-programme/reports/world-malaria...
-
- Gutman J, Kovacs S, Dorsey G, Stergachis A, Ter Kuile FO. 2017. Safety, tolerability, and efficacy of repeated doses of dihydroartemisinin-piperaquine for prevention and treatment of malaria: a systematic review and meta-analysis. Lancet Infect Dis 17:184–193. doi:10.1016/S1473-3099(16)30378-4 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

