Multimodal assessment of mitochondrial function in Parkinson's disease
- PMID: 38059801
- PMCID: PMC10766247
- DOI: 10.1093/brain/awad364
Multimodal assessment of mitochondrial function in Parkinson's disease
Abstract
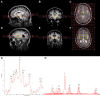
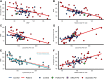
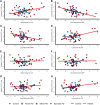
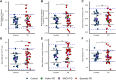
The heterogenous aetiology of Parkinson's disease is increasingly recognized; both mitochondrial and lysosomal dysfunction have been implicated. Powerful, clinically applicable tools are required to enable mechanistic stratification for future precision medicine approaches. The aim of this study was to characterize bioenergetic dysfunction in Parkinson's disease by applying a multimodal approach, combining standardized clinical assessment with midbrain and putaminal 31-phosphorus magnetic resonance spectroscopy (31P-MRS) and deep phenotyping of mitochondrial and lysosomal function in peripheral tissue in patients with recent-onset Parkinson's disease and control subjects. Sixty participants (35 patients with Parkinson's disease and 25 healthy controls) underwent 31P-MRS for quantification of energy-rich metabolites [ATP, inorganic phosphate (Pi) and phosphocreatine] in putamen and midbrain. In parallel, skin biopsies were obtained from all research participants to establish fibroblast cell lines for subsequent quantification of total intracellular ATP and mitochondrial membrane potential (MMP) as well as mitochondrial and lysosomal morphology, using high content live cell imaging. Lower MMP correlated with higher intracellular ATP (r = -0.55, P = 0.0016), higher mitochondrial counts (r = -0.72, P < 0.0001) and higher lysosomal counts (r = -0.62, P = 0.0002) in Parkinson's disease patient-derived fibroblasts only, consistent with impaired mitophagy and mitochondrial uncoupling. 31P-MRS-derived posterior putaminal Pi/ATP ratio variance was considerably greater in Parkinson's disease than in healthy controls (F-tests, P = 0.0036). Furthermore, elevated 31P-MRS-derived putaminal, but not midbrain Pi/ATP ratios (indicative of impaired oxidative phosphorylation) correlated with both greater mitochondrial (r = 0.37, P = 0.0319) and lysosomal counts (r = 0.48, P = 0.0044) as well as lower MMP in both short (r = -0.52, P = 0.0016) and long (r = -0.47, P = 0.0052) mitochondria in Parkinson's disease. Higher 31P-MRS midbrain phosphocreatine correlated with greater risk of rapid disease progression (r = 0.47, P = 0.0384). Our data suggest that impaired oxidative phosphorylation in the striatal dopaminergic nerve terminals exceeds mitochondrial dysfunction in the midbrain of patients with early Parkinson's disease. Our data further support the hypothesis of a prominent link between impaired mitophagy and impaired striatal energy homeostasis as a key event in early Parkinson's disease.
Keywords: 31phosphorus magnetic resonance spectroscopy; Parkinson’s disease; disease stratification; fibroblasts; mitochondria.
© The Author(s) 2023. Published by Oxford University Press on behalf of the Guarantors of Brain.
Conflict of interest statement
The authors report no competing interests.
Figures

References
-
- Dorsey ER, Constantinescu R, Thompson JP, et al. . Projected number of people with Parkinson disease in the most populous nations, 2005 through 2030. Neurology. 2007;68:384–386. - PubMed
-
- Vijiaratnam N, Simuni T, Bandmann O, Morris HR, Foltynie T. Progress towards therapies for disease modification in Parkinson's disease. Lancet Neurol. 2021;20:559–572. - PubMed
-
- Schapira AH, Cooper JM, Dexter D, Clark JB, Jenner P, Marsden CD. Mitochondrial complex I deficiency in Parkinson’s disease. J Neurochem. 1990;54:823–827. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

