Hemorrhage-activated NRF2 in tumor-associated macrophages drives cancer growth, invasion, and immunotherapy resistance
- PMID: 38060331
- PMCID: PMC10849758
- DOI: 10.1172/JCI174528
Hemorrhage-activated NRF2 in tumor-associated macrophages drives cancer growth, invasion, and immunotherapy resistance
Abstract
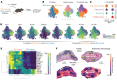
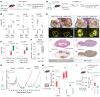
Microscopic hemorrhage is a common aspect of cancers, yet its potential role as an independent factor influencing both cancer progression and therapeutic response is largely ignored. Recognizing the essential function of macrophages in red blood cell disposal, we explored a pathway that connects intratumoral hemorrhage with the formation of cancer-promoting tumor-associated macrophages (TAMs). Using spatial transcriptomics, we found that NRF2-activated myeloid cells possessing characteristics of procancerous TAMs tend to cluster in perinecrotic hemorrhagic tumor regions. These cells resembled antiinflammatory erythrophagocytic macrophages. We identified heme, a red blood cell metabolite, as a pivotal microenvironmental factor steering macrophages toward protumorigenic activities. Single-cell RNA-Seq and functional assays of TAMs in 3D cell culture spheroids revealed how elevated intracellular heme signals via the transcription factor NRF2 to induce cancer-promoting TAMs. These TAMs stabilized epithelial-mesenchymal transition, enhancing cancer invasiveness and metastatic potential. Additionally, NRF2-activated macrophages exhibited resistance to reprogramming by IFN-γ and anti-CD40 antibodies, reducing their tumoricidal capacity. Furthermore, MC38 colon adenocarcinoma-bearing mice with NRF2 constitutively activated in leukocytes were resistant to anti-CD40 immunotherapy. Overall, our findings emphasize hemorrhage-activated NRF2 in TAMs as a driver of cancer progression, suggesting that targeting this pathway could offer new strategies to enhance cancer immunity and overcome therapy resistance.
Keywords: Cancer; Inflammation; Innate immunity; Macrophages; Oncology.
Conflict of interest statement
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

