An inducible Cd79b mutation confers ibrutinib sensitivity in mouse models of Myd88-driven diffuse large B-cell lymphoma
- PMID: 38060829
- PMCID: PMC10907402
- DOI: 10.1182/bloodadvances.2023011213
An inducible Cd79b mutation confers ibrutinib sensitivity in mouse models of Myd88-driven diffuse large B-cell lymphoma
Abstract
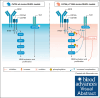
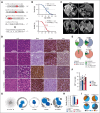
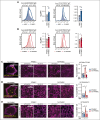
Diffuse large B-cell lymphoma (DLBCL) is the most common aggressive lymphoma and constitutes a highly heterogenous disease. Recent comprehensive genomic profiling revealed the identity of numerous molecularly defined DLBCL subtypes, including a cluster which is characterized by recurrent aberrations in MYD88, CD79B, and BCL2, as well as various lesions promoting a block in plasma cell differentiation, including PRDM1, TBL1XR1, and SPIB. Here, we generated a series of autochthonous mouse models to mimic this DLBCL cluster and specifically focused on the impact of Cd79b mutations in this setting. We show that canonical Cd79b immunoreceptor tyrosine-based activation motif (ITAM) mutations do not accelerate Myd88- and BCL2-driven lymphomagenesis. Cd79b-mutant murine DLBCL were enriched for IgM surface expression, reminiscent of their human counterparts. Moreover, Cd79b-mutant lymphomas displayed a robust formation of cytoplasmic signaling complexes involving MYD88, CD79B, MALT1, and BTK. These complexes were disrupted upon pharmacological BTK inhibition. The BTK inhibitor-mediated disruption of these signaling complexes translated into a selective ibrutinib sensitivity of lymphomas harboring combined Cd79b and Myd88 mutations. Altogether, this in-depth cross-species comparison provides a framework for the development of molecularly targeted therapeutic intervention strategies in DLBCL.
© 2024 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: H.C.R. received consulting and lecture fees from AbbVie, AstraZeneca, Roche, Janssen-Cilag, Novartis, Vertex, and Merck; research funding from Gilead and AstraZeneca; and is a cofounder of CDL Therapeutics GmbH. B.v.T. is an adviser or consultant for Allogene, BMS/Celgene, Cerus, Incyte, IQVIA, Gilead Kite, Miltenyi, Novartis, Noscendo, Pentixapharm, Roche, Amgen, Pfizer, Takeda, Merck Sharp & Dohme, and Gilead Kite; has received honoraria from AstraZeneca, BMS, Incyte, Novartis, Roche Pharma AG, Takeda, and Merck Sharp & Dohme; reports research funding from Novartis (institutional), Merck Sharp & Dohme (institutional), and Takeda (institutional); and travel support from AbbVie, AstraZeneca, Gilead Kite, Merck Sharp & Dohme, Roche, Takeda, and Novartis. The remaining authors declare no competing financial interests.
Figures


References
-
- Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503–511. - PubMed
-
- Rosenwald A, Wright G, Chan WC, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346(25):1937–1947. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

