Outward-oriented sites within clustered CTCF boundaries are key for intra-TAD chromatin interactions and gene regulation
- PMID: 38062010
- PMCID: PMC10703910
- DOI: 10.1038/s41467-023-43849-0
Outward-oriented sites within clustered CTCF boundaries are key for intra-TAD chromatin interactions and gene regulation
Abstract
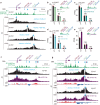
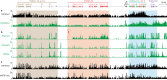
CTCF plays an important role in 3D genome organization by adjusting the strength of chromatin insulation at TAD boundaries, where clustered CBS (CTCF-binding site) elements are often arranged in a tandem array with a complex divergent or convergent orientation. Here, using Pcdh and HOXD loci as a paradigm, we look into the clustered CTCF TAD boundaries and find that, counterintuitively, outward-oriented CBS elements are crucial for inward enhancer-promoter interactions as well as for gene regulation. Specifically, by combinatorial deletions of a series of putative enhancer elements in mice in vivo or CBS elements in cultured cells in vitro, in conjunction with chromosome conformation capture and RNA-seq analyses, we show that deletions of outward-oriented CBS elements weaken the strength of long-distance intra-TAD promoter-enhancer interactions and enhancer activation of target genes. Our data highlight the crucial role of outward-oriented CBS elements within the clustered CTCF TAD boundaries in developmental gene regulation and have interesting implications on the organization principles of clustered CTCF sites within TAD boundaries.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

