SEMA6A drives GnRH neuron-dependent puberty onset by tuning median eminence vascular permeability
- PMID: 38062045
- PMCID: PMC10703890
- DOI: 10.1038/s41467-023-43820-z
SEMA6A drives GnRH neuron-dependent puberty onset by tuning median eminence vascular permeability
Abstract
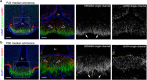
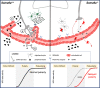
Innervation of the hypothalamic median eminence by Gonadotropin-Releasing Hormone (GnRH) neurons is vital to ensure puberty onset and successful reproduction. However, the molecular and cellular mechanisms underlying median eminence development and pubertal timing are incompletely understood. Here we show that Semaphorin-6A is strongly expressed by median eminence-resident oligodendrocytes positioned adjacent to GnRH neuron projections and fenestrated capillaries, and that Semaphorin-6A is required for GnRH neuron innervation and puberty onset. In vitro and in vivo experiments reveal an unexpected function for Semaphorin-6A, via its receptor Plexin-A2, in the control of median eminence vascular permeability to maintain neuroendocrine homeostasis. To support the significance of these findings in humans, we identify patients with delayed puberty carrying a novel pathogenic variant of SEMA6A. In all, our data reveal a role for Semaphorin-6A in regulating GnRH neuron patterning by tuning the median eminence vascular barrier and thereby controlling puberty onset.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







References
-
- Prevot V, et al. Definitive evidence for the existence of morphological plasticity in the external zone of the median eminence during the rat estrous cycle: implication of neuro-glio-endothelial interactions in gonadotropin-releasing hormone release. Neuroscience. 1999;94:809–819. doi: 10.1016/S0306-4522(99)00383-8. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

