Endometrial cancer PDX-derived organoids (PDXOs) and PDXs with FGFR2c isoform expression are sensitive to FGFR inhibition
- PMID: 38062117
- PMCID: PMC10703877
- DOI: 10.1038/s41698-023-00478-6
Endometrial cancer PDX-derived organoids (PDXOs) and PDXs with FGFR2c isoform expression are sensitive to FGFR inhibition
Abstract
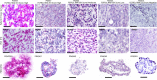
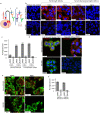
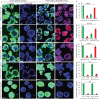
Endometrial cancer (EC) patients with metastatic/recurrent disease have limited treatment options and poor survival outcomes. Recently, we discovered the FGFR2c splice isoform is associated with poor prognosis in EC patients. Here we report the establishment of 16 EC patient-derived xenografts (PDX)-derived organoids (PDXOs) with or without FGFR2c expression. In vitro treatment of 5 EC PDXOs with BGJ398 showed significant cell death in 3 models with FGFR2c expression. PDXs with high/moderate FGFR2c expression showed significant tumour growth inhibition (TGI) following 21-day treatment with FGFR inhibitors (BGJ398 or pemigatinib) and significantly prolonged survival in 4/5 models. Pemigatinib + cisplatin combination therapy (n = 5) resulted in significant TGI and prolonged survival in one of two p53abn PDXs. All five models treated with cisplatin alone showed de novo resistance and no survival benefit. Seven-day treatment with BGJ398 revealed a significant reduction in angiogenesis and CD206 + M2 macrophages. These data collectively support the evaluation of FGFR inhibitors in a clinical trial.
© 2023. The Author(s).
Conflict of interest statement
Asmerom Sengal and Pamela Pollock received funding support from Incyte through an investigator-initiated study (IIS) grant scheme. The rest of the authors declare that there are no competing interests.
Figures






References
-
- Sung H, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: A Cancer J. Clin. 2021;71:209–249. - PubMed
LinkOut - more resources
Full Text Sources

