Attention-based neural networks for clinical prediction modelling on electronic health records
- PMID: 38062352
- PMCID: PMC10701944
- DOI: 10.1186/s12874-023-02112-2
Attention-based neural networks for clinical prediction modelling on electronic health records
Abstract
Background: Deep learning models have had a lot of success in various fields. However, on structured data they have struggled. Here we apply four state-of-the-art supervised deep learning models using the attention mechanism and compare against logistic regression and XGBoost using discrimination, calibration and clinical utility.
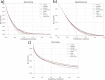
Methods: We develop the models using a general practitioners database. We implement a recurrent neural network, a transformer with and without reverse distillation and a graph neural network. We measure discrimination using the area under the receiver operating characteristic curve (AUC) and the area under the precision recall curve (AUPRC). We assess smooth calibration using restricted cubic splines and clinical utility with decision curve analysis.
Results: Our results show that deep learning approaches can improve discrimination up to 2.5% points AUC and 7.4% points AUPRC. However, on average the baselines are competitive. Most models are similarly calibrated as the baselines except for the graph neural network. The transformer using reverse distillation shows the best performance in clinical utility on two out of three prediction problems over most of the prediction thresholds.
Conclusion: In this study, we evaluated various approaches in supervised learning using neural networks and attention. Here we do a rigorous comparison, not only looking at discrimination but also calibration and clinical utility. There is value in using deep learning models on electronic health record data since it can improve discrimination and clinical utility while providing good calibration. However, good baseline methods are still competitive.
Keywords: Clinical prediction models; Deep learning; Electronic health records.
© 2023. The Author(s).
Conflict of interest statement
DS was a consultant for and has equity in CureAI and ASAPP and has received compensation from speaking at Genentech. DS has a grant from Takeda. EF and PR work for a research group who received unconditional research grants from Boehringer-Ingelheim, GSK, Janssen Research & Development, Novartis, Pfizer, Yamanouchi, Servier. None of these grants result in a conflict of interest to the content of this paper.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

