Comparative genomic analysis of clinical Enterococcus faecalis distinguishes strains isolated from the bladder
- PMID: 38062354
- PMCID: PMC10701997
- DOI: 10.1186/s12864-023-09818-z
Comparative genomic analysis of clinical Enterococcus faecalis distinguishes strains isolated from the bladder
Abstract
Background: Enterococcus faecalis is the most commonly isolated enterococcal species in clinical infection. This bacterium is notorious for its ability to share genetic content within and outside of its species. With this increased proficiency for horizontal gene transfer, tremendous genomic diversity within this species has been identified. Many researchers have hypothesized E. faecalis exhibits niche adaptation to establish infections or colonize various parts of the human body. Here, we hypothesize that E. faecalis strains isolated from the human bladder will carry unique genomic content compared to clinical strains isolated from other sources.
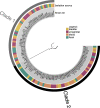
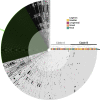
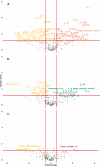
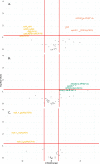
Results: This analysis includes comparison of 111 E. faecalis genomes isolated from bladder, urogenital, blood, and fecal samples. Phylogenomic comparison shows no association between isolation source and lineage; however, accessory genome comparison differentiates blood and bladder genomes. Further gene enrichment analysis identifies gene functions, virulence factors, antibiotic resistance genes, and plasmid-associated genes that are enriched or rare in bladder genomes compared to urogenital, blood, and fecal genomes. Using these findings as training data and 682 publicly available genomes as test data, machine learning classifiers successfully distinguished between bladder and non-bladder strains with high accuracy. Genes identified as important for this differentiation were often related to transposable elements and phage, including 3 prophage species found almost exclusively in bladder and urogenital genomes.
Conclusions: E. faecalis strains isolated from the bladder contain unique genomic content when compared to strains isolated from other body sites. This genomic diversity is most likely due to horizontal gene transfer, as evidenced by lack of phylogenomic clustering and enrichment of transposable elements and prophages. Investigation into how these enriched genes influence host-microbe interactions may elucidate gene functions required for successful bladder colonization and disease establishment.
Keywords: Bladder microbiome; Comparative pangenomics; Enterococcus faecalis genomics; Niche adaptation.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Comparative genomics of Enterococcus spp. isolated from bovine feces.BMC Microbiol. 2017 Mar 8;17(1):52. doi: 10.1186/s12866-017-0962-1. BMC Microbiol. 2017. PMID: 28270110 Free PMC article.
-
Investigating the mobilome in clinically important lineages of Enterococcus faecium and Enterococcus faecalis.BMC Genomics. 2015 Apr 10;16:282. doi: 10.1186/s12864-015-1407-6. BMC Genomics. 2015. PMID: 25885771 Free PMC article.
-
Nonclinical and clinical Enterococcus faecium strains, but not Enterococcus faecalis strains, have distinct structural and functional genomic features.Appl Environ Microbiol. 2014 Jan;80(1):154-65. doi: 10.1128/AEM.03108-13. Epub 2013 Oct 18. Appl Environ Microbiol. 2014. PMID: 24141120 Free PMC article.
-
Enterococcal Genomics.2014 Feb 8. In: Gilmore MS, Clewell DB, Ike Y, Shankar N, editors. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection [Internet]. Boston: Massachusetts Eye and Ear Infirmary; 2014–. 2014 Feb 8. In: Gilmore MS, Clewell DB, Ike Y, Shankar N, editors. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection [Internet]. Boston: Massachusetts Eye and Ear Infirmary; 2014–. PMID: 24649511 Free Books & Documents. Review.
-
Review of virulence factors of enterococcus: an emerging nosocomial pathogen.Indian J Med Microbiol. 2009 Oct-Dec;27(4):301-5. doi: 10.4103/0255-0857.55437. Indian J Med Microbiol. 2009. PMID: 19736397 Review.
Cited by
-
Insights into ecology, pathogenesis, and biofilm formation of Enterococcus faecalis from functional genomics.Microbiol Mol Biol Rev. 2025 Mar 27;89(1):e0008123. doi: 10.1128/mmbr.00081-23. Epub 2024 Dec 23. Microbiol Mol Biol Rev. 2025. PMID: 39714182 Review.
-
Genomic diversity, antibiotic resistance, and virulence in South African Enterococcus faecalis and Enterococcus lactis isolates.World J Microbiol Biotechnol. 2024 Aug 5;40(10):289. doi: 10.1007/s11274-024-04098-5. World J Microbiol Biotechnol. 2024. PMID: 39102038 Free PMC article.
-
Comprehensive analysis of Enterococcus spp. from two European healthy infant cohorts shows stable genomic traits including antimicrobial resistance (AMR).Gut Microbes. 2025 Dec;17(1):2516699. doi: 10.1080/19490976.2025.2516699. Epub 2025 Jun 16. Gut Microbes. 2025. PMID: 40518985 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

