Tracking DNA-based antigen-specific T cell receptors during progression to type 1 diabetes
- PMID: 38064552
- PMCID: PMC10708189
- DOI: 10.1126/sciadv.adj6975
Tracking DNA-based antigen-specific T cell receptors during progression to type 1 diabetes
Abstract
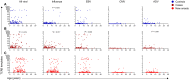
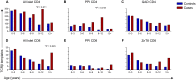
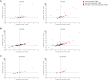
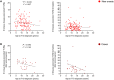
T cells targeting self-proteins are important mediators in autoimmune diseases. T cells express unique cell-surface receptors (TCRs) that recognize peptides presented by major histocompatibility molecules. TCRs have been identified from blood and pancreatic islets of individuals with type 1 diabetes (T1D). Here, we tracked ~1700 known antigen-specific TCR sequences, islet antigen or viral reactive, in bulk TCRβ sequencing from longitudinal blood DNA samples in at-risk cases who progressed to T1D, age/sex/human leukocyte antigen-matched controls, and a new-onset T1D cohort. Shared and frequent antigen-specific TCRβ sequences were identified in all three cohorts, and viral sequences were present across all ages. Islet sequences had different patterns of accumulation based upon antigen specificity in the at-risk cases. Furthermore, 73 islet-antigen TCRβ sequences were present in higher frequencies and numbers in T1D samples relative to controls. The total number of these disease-associated TCRβ sequences inversely correlated with age at clinical diagnosis, indicating the potential to use disease-relevant TCR sequences as biomarkers in autoimmune disorders.
Figures


References
-
- Emerson R. O., DeWitt W. S., Vignali M., Gravley J., Hu J. K., Osborne E. J., Desmarais C., Klinger M., Carlson C. S., Hansen J. A., Rieder M., Robins H. S., Immunosequencing identifies signatures of cytomegalovirus exposure history and HLA-mediated effects on the T cell repertoire. Nat. Genet. 49, 659–665 (2017). - PubMed
-
- Elyanow R., Snyder T. M., Dalai S. C., Gittelman R. M., Boonyaratanakornkit J., Wald A., Selke S., Wener M. H., Morishima C., Greninger A. L., Gale M., Hsiang T.-Y., Jing L., Holbrook M. R., Kaplan I. M., Zahid H. J., May D. H., Carlson J. M., Baldo L., Manley T., Robins H. S., Koelle D. M., T cell receptor sequencing identifies prior SARS-CoV-2 infection and correlates with neutralizing antibodies and disease severity. JCI insight 7, e150070 (2022). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

