SON is an essential m6A target for hematopoietic stem cell fate
- PMID: 38065069
- PMCID: PMC10752439
- DOI: 10.1016/j.stem.2023.11.006
SON is an essential m6A target for hematopoietic stem cell fate
Abstract
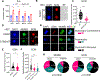
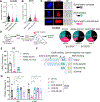
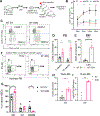
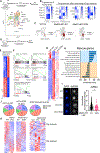
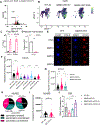
Stem cells regulate their self-renewal and differentiation fate outcomes through both symmetric and asymmetric divisions. m6A RNA methylation controls symmetric commitment and inflammation of hematopoietic stem cells (HSCs) through unknown mechanisms. Here, we demonstrate that the nuclear speckle protein SON is an essential m6A target required for murine HSC self-renewal, symmetric commitment, and inflammation control. Global profiling of m6A identified that m6A mRNA methylation of Son increases during HSC commitment. Upon m6A depletion, Son mRNA increases, but its protein is depleted. Reintroduction of SON rescues defects in HSC symmetric commitment divisions and engraftment. Conversely, Son deletion results in a loss of HSC fitness, while overexpression of SON improves mouse and human HSC engraftment potential by increasing quiescence. Mechanistically, we found that SON rescues MYC and suppresses the METTL3-HSC inflammatory gene expression program, including CCL5, through transcriptional regulation. Thus, our findings define a m6A-SON-CCL5 axis that controls inflammation and HSC fate.
Keywords: RNA binding proteins; RNA methylation; RNA modifications; SON; cell fate; differentiation; hematopoietic stem cells; inflammation; nuclear speckles; stem cells.
Copyright © 2023 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests M.G.K. is a SAB member of 858 Therapeutics and received honorarium from Kumquat, AstraZeneca, and consulting at Transition Bio. D.A.L. has served as a consultant for AbbVie, AstraZeneca, and Illumina and is on the Scientific Advisory Board of Mission Bio, Pangea, Alethiomics, and C2i Genomics; D.A.L. has received prior research funding from BMS, 10x Genomics, Ultima Genomics, and Illumina unrelated to the current manuscript.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
- R01 HG013328/HG/NHGRI NIH HHS/United States
- R01 CA236911/CA/NCI NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- R01 HL157387/HL/NHLBI NIH HHS/United States
- R01 HL168659/HL/NHLBI NIH HHS/United States
- R01 CA193842/CA/NCI NIH HHS/United States
- R01 CA283578/CA/NCI NIH HHS/United States
- K99 DK128602/DK/NIDDK NIH HHS/United States
- R01 CA225231/CA/NCI NIH HHS/United States
- R01 CA274249/CA/NCI NIH HHS/United States
- R01 CA186702/CA/NCI NIH HHS/United States
- R01 DK101989/DK/NIDDK NIH HHS/United States
- P50 CA254838/CA/NCI NIH HHS/United States
- R01 HL135564/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

